> Dataset: GSE140662
|
|
HeatMap of cell type fractions in all samples
|
Lollipop of FGF3 expression and immune infiltrating cell level correlation result in HPV+ samples
|
FGF3 expression: Low -> High
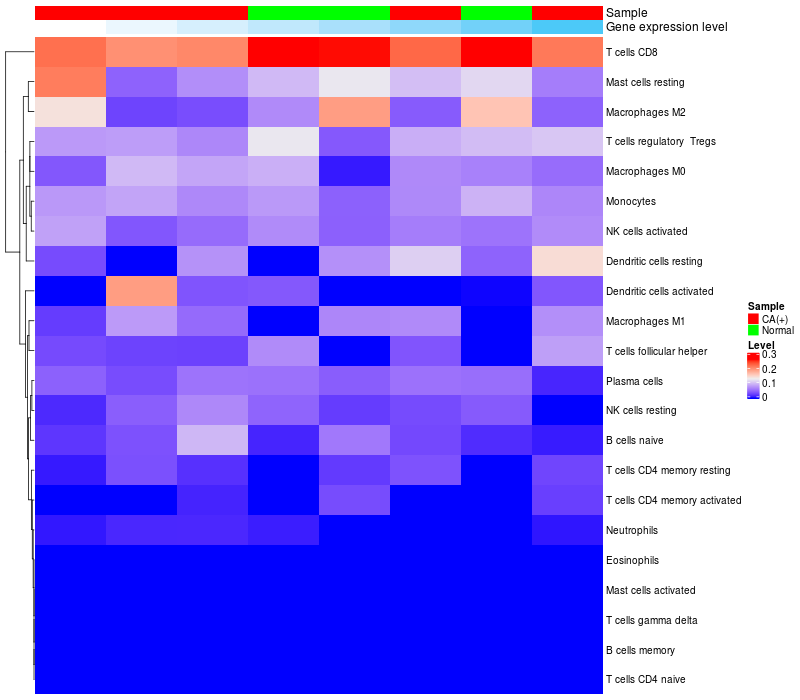
|
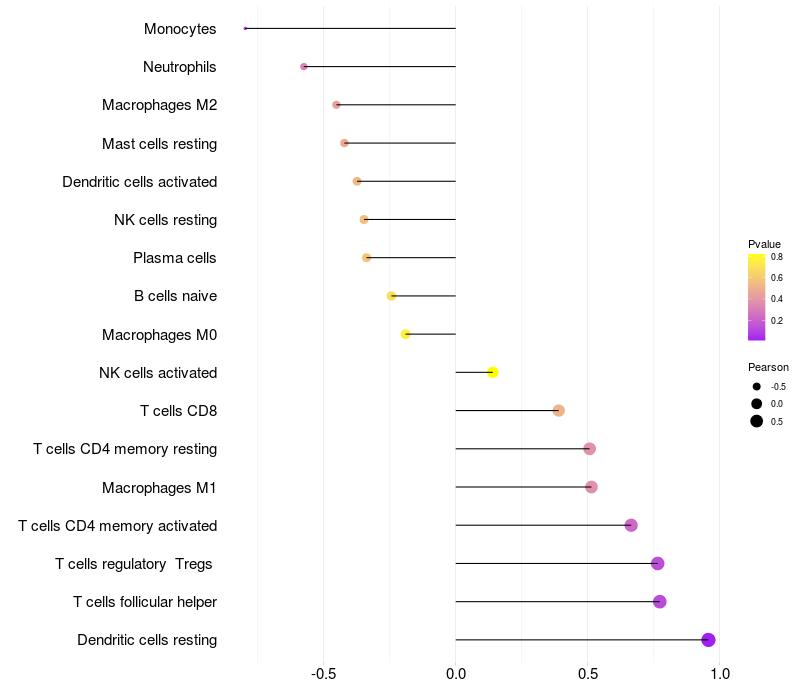
|
|
> Dataset: GSE65858
|
|
HeatMap of cell type fractions in all samples
|
Lollipop of FGF3 expression and immune infiltrating cell level correlation result in HPV+ samples
|
FGF3 expression: Low -> High
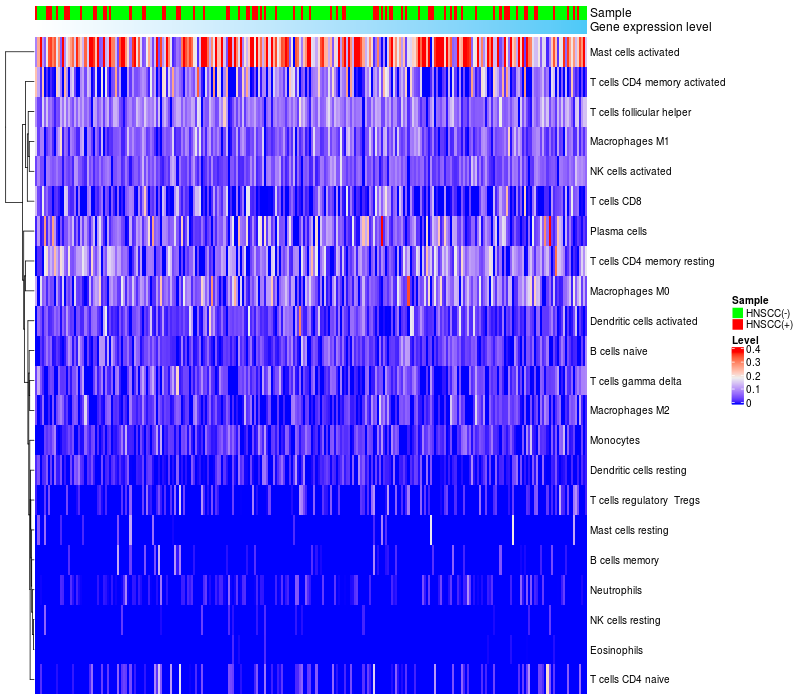
|
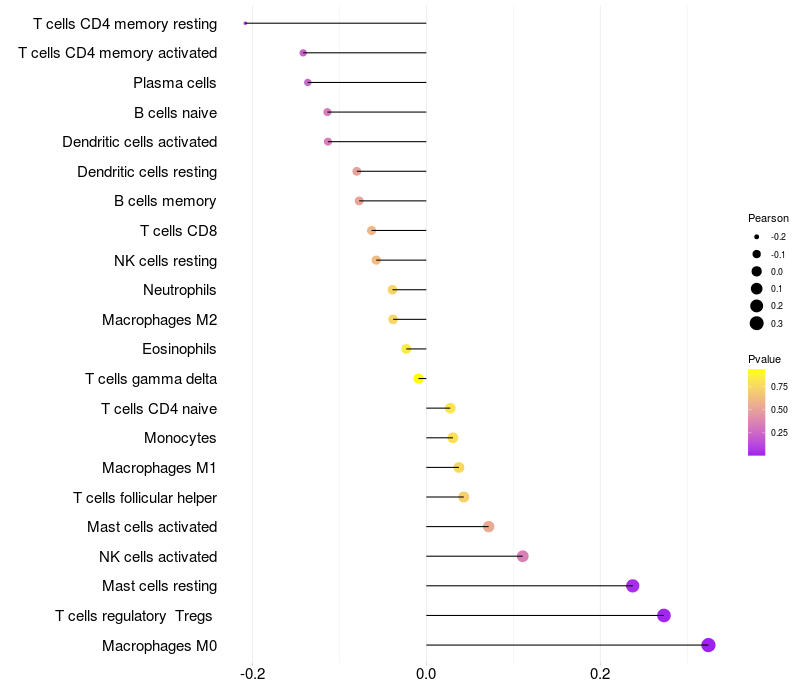
|
|
> Dataset: GSE55546
|
|
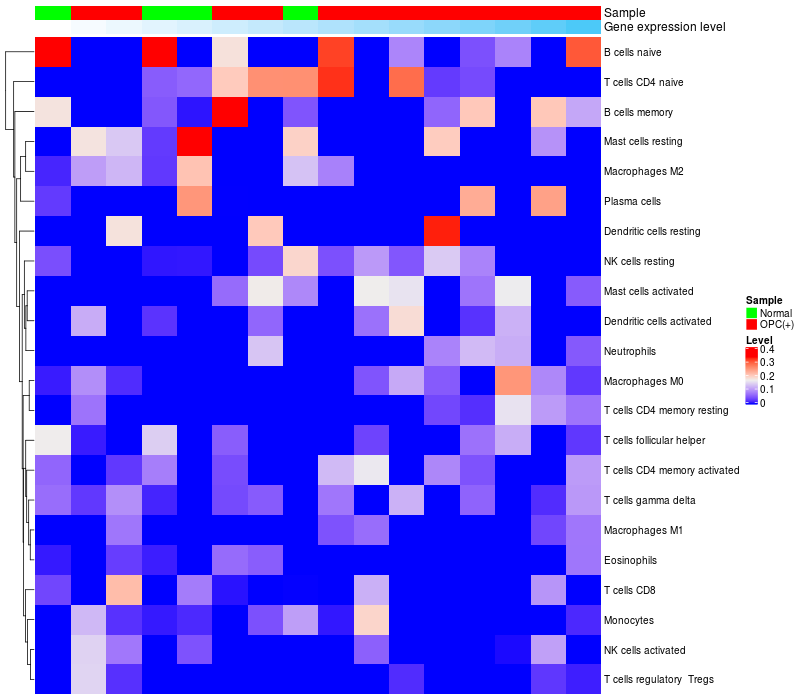
HeatMap of cell type fractions in all samples
|
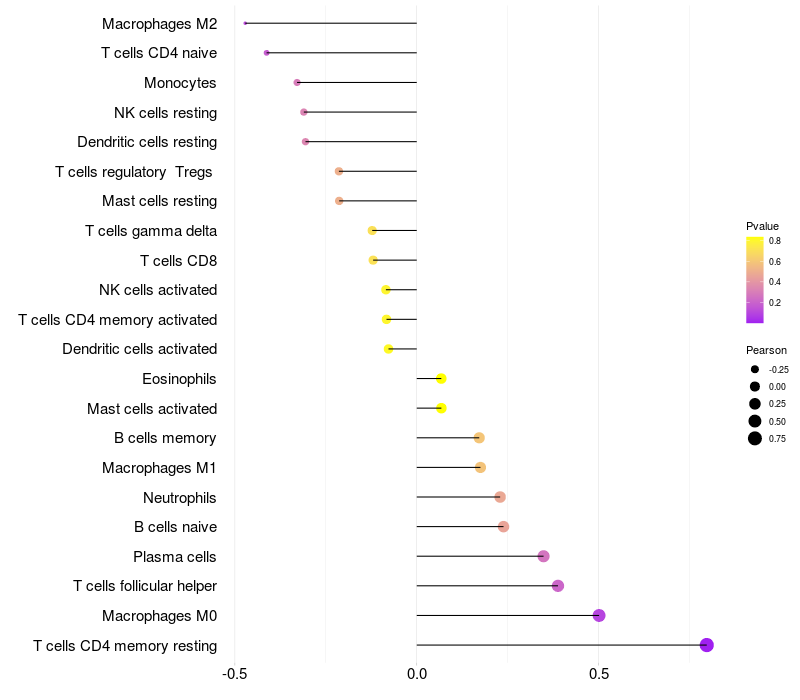
Lollipop of FGF3 expression and immune infiltrating cell level correlation result in HPV+ samples
|
FGF3 expression: Low -> High
|
|
|
> Dataset: GSE55544
|
|
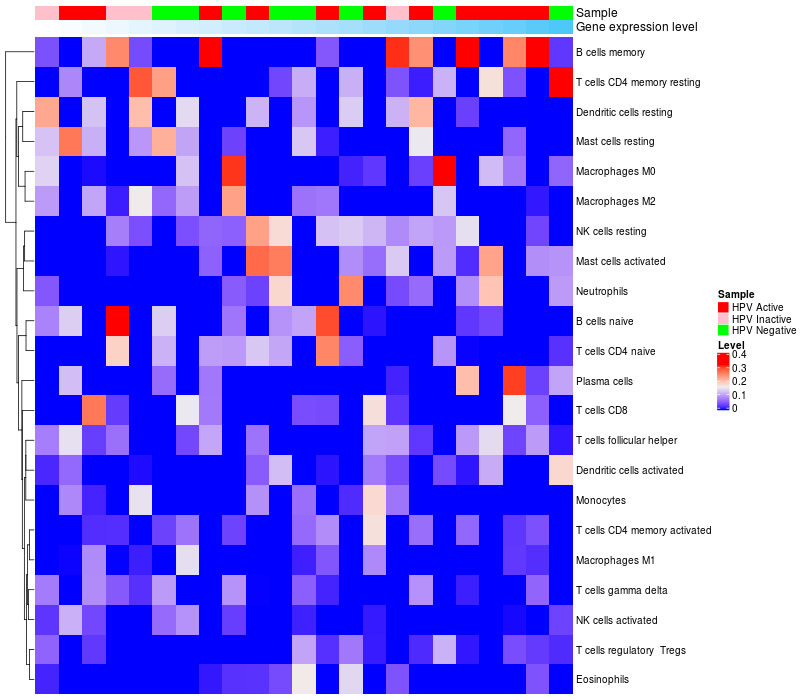
HeatMap of cell type fractions in all samples
|
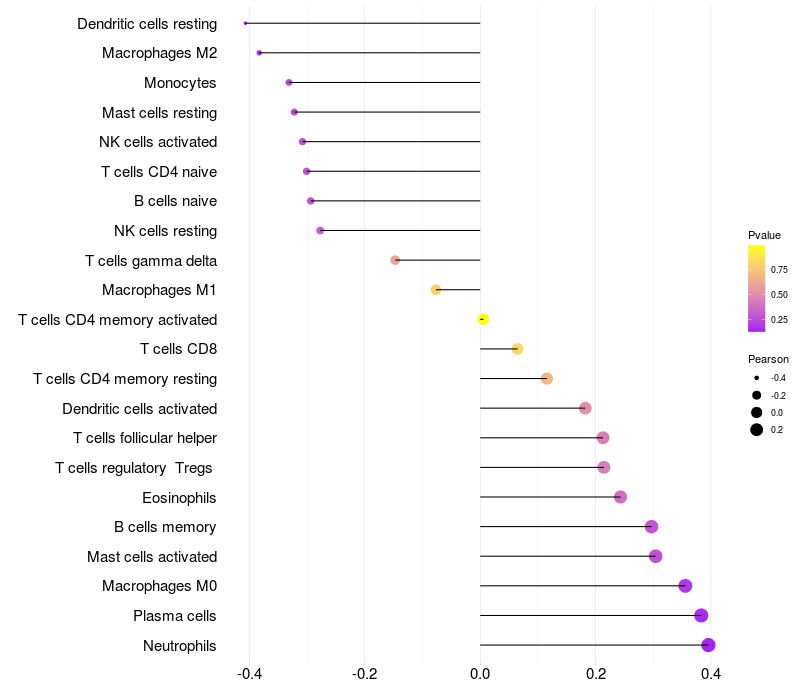
Lollipop of FGF3 expression and immune infiltrating cell level correlation result in HPV+ samples
|
FGF3 expression: Low -> High
|
|
|
> Dataset: GSE55542
|
|
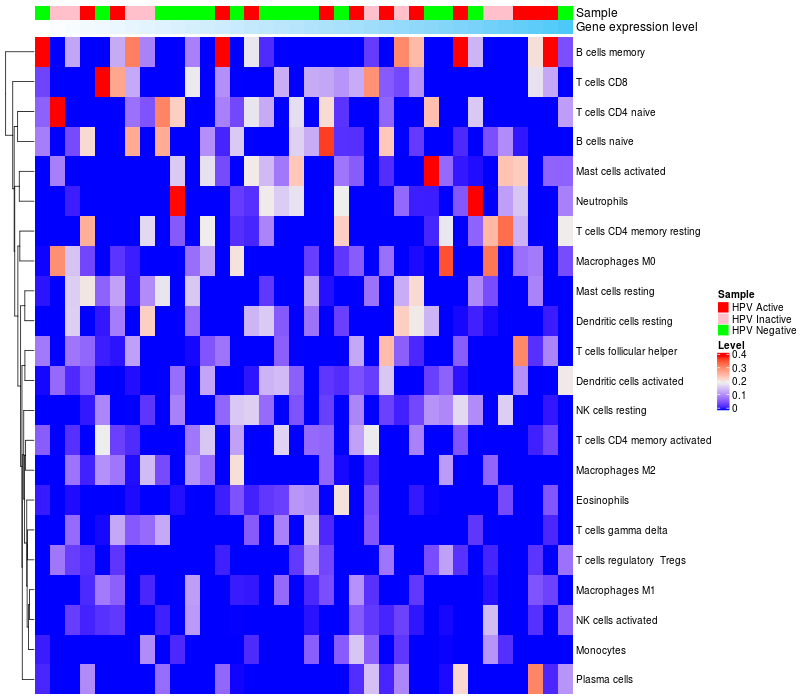
HeatMap of cell type fractions in all samples
|
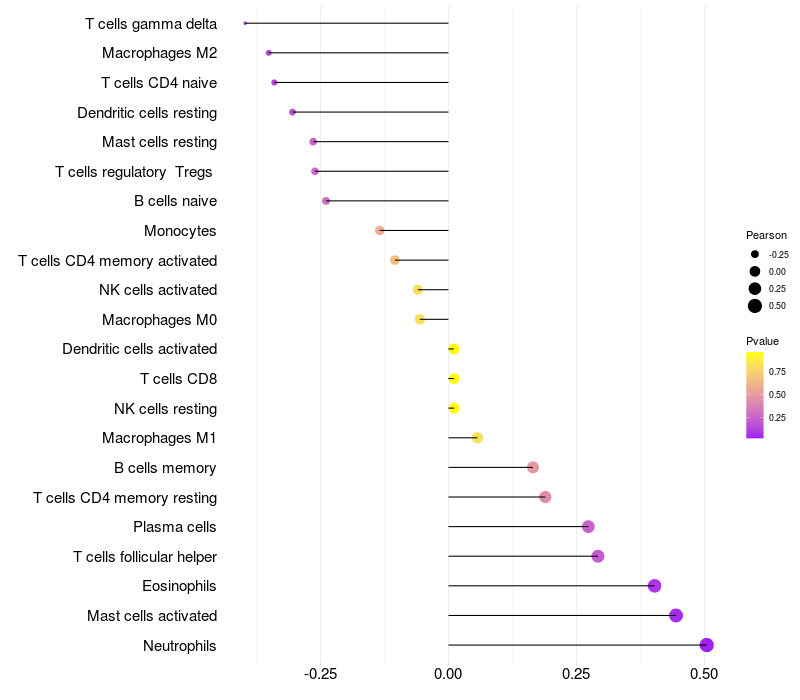
Lollipop of FGF3 expression and immune infiltrating cell level correlation result in HPV+ samples
|
FGF3 expression: Low -> High
|
|
|
> Dataset: GSE138080
|
|
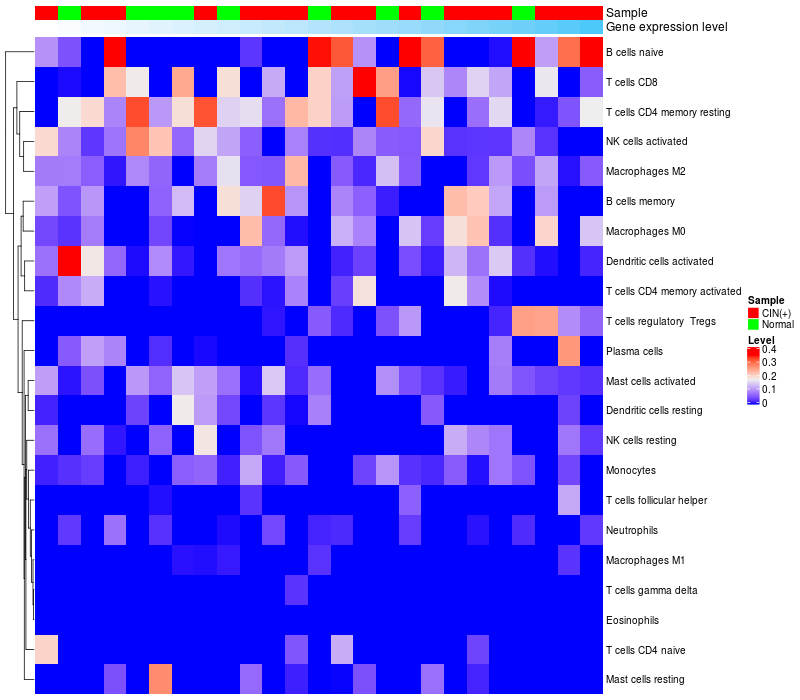
HeatMap of cell type fractions in all samples
|
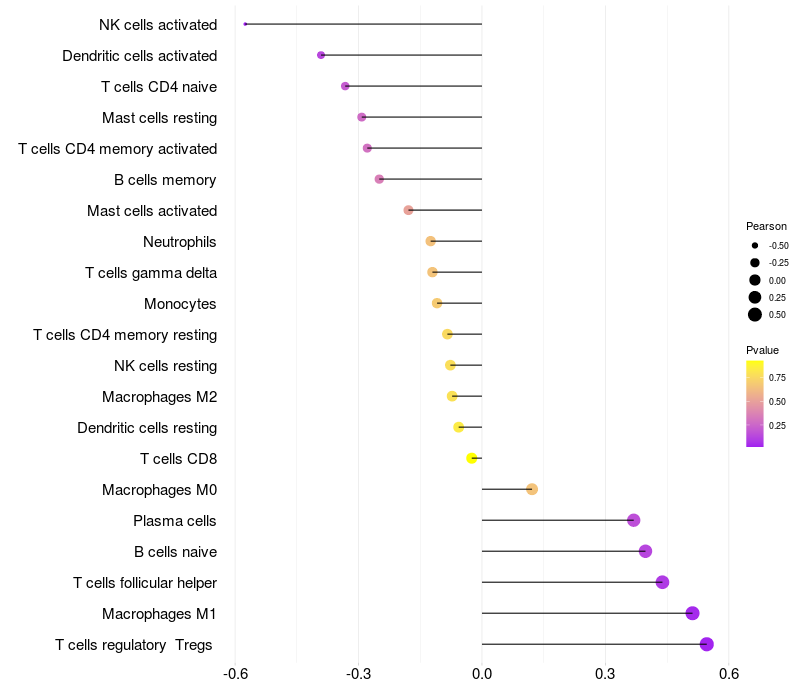
Lollipop of FGF3 expression and immune infiltrating cell level correlation result in HPV+ samples
|
FGF3 expression: Low -> High
|
|
|
> Dataset: GSE51993
|
|
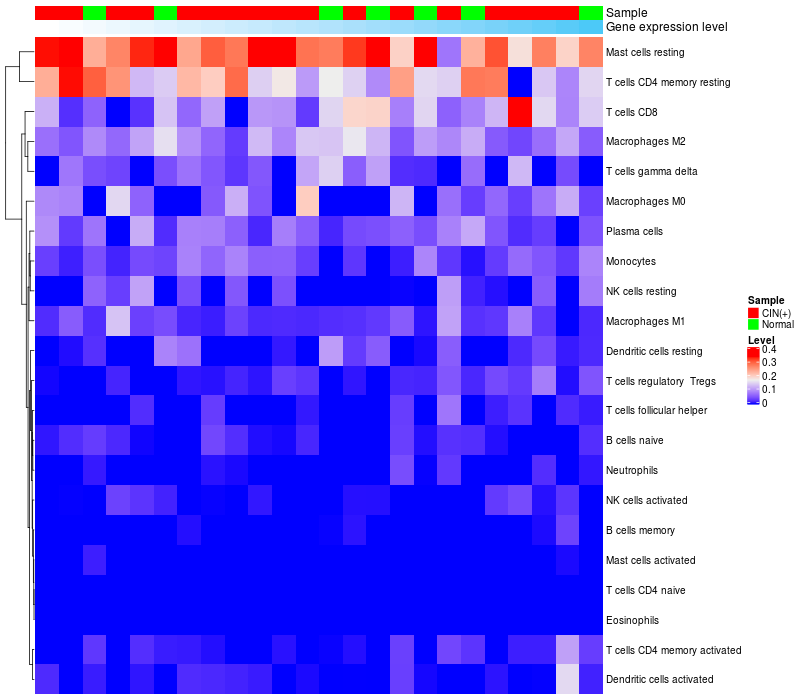
HeatMap of cell type fractions in all samples
|
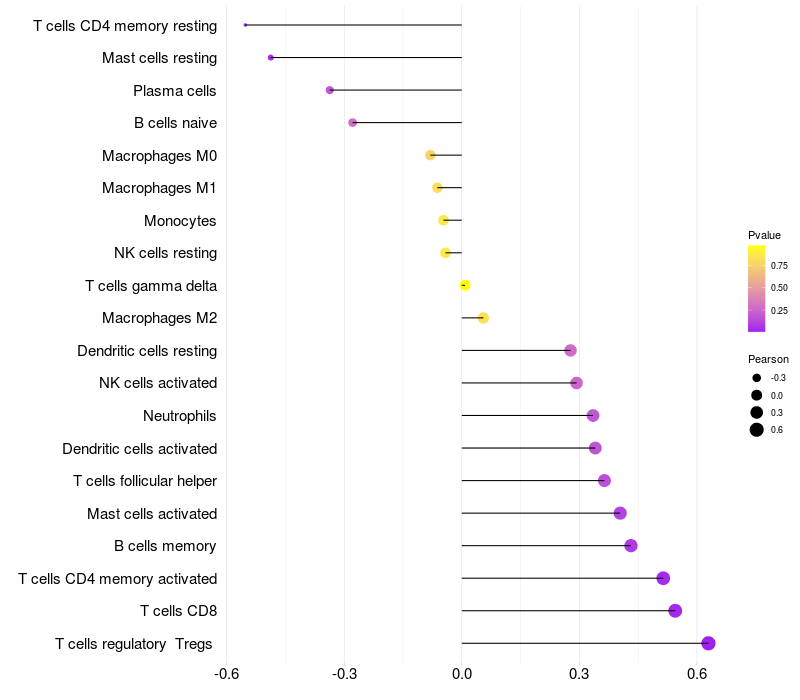
Lollipop of FGF3 expression and immune infiltrating cell level correlation result in HPV+ samples
|
FGF3 expression: Low -> High
|
|
|
> Dataset: HNSC
|
|
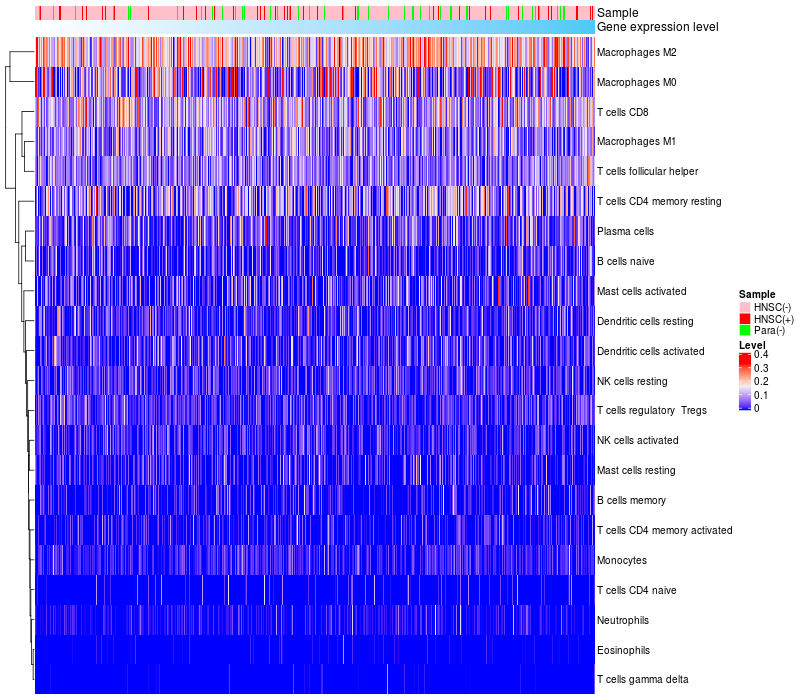
HeatMap of cell type fractions in all samples
|
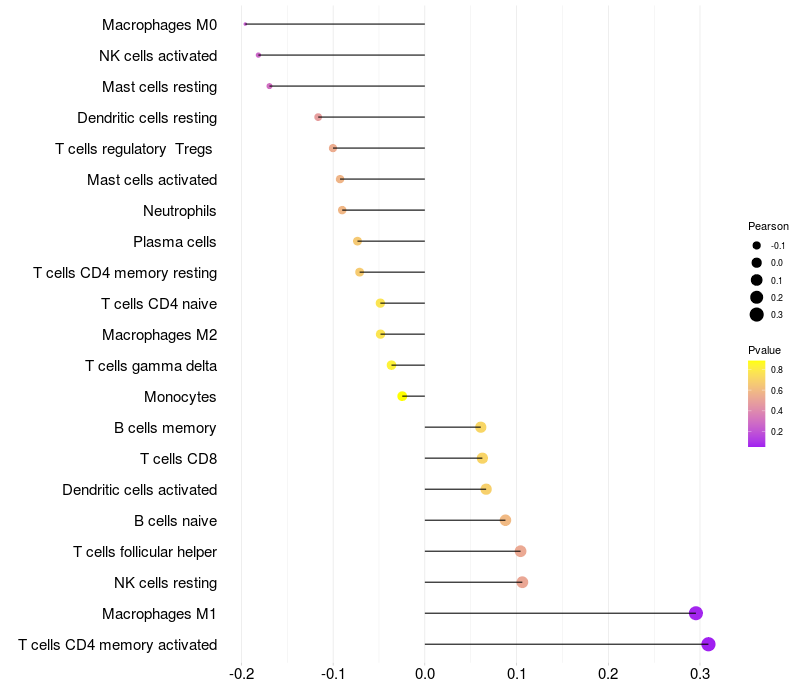
Lollipop of FGF3 expression and immune infiltrating cell level correlation result in HPV+ samples
|
FGF3 expression: Low -> High
|
|
|
 HPV Target gene Detail Information
HPV Target gene Detail Information