Overview
Welcome to ViMIC (A Database of Human Disease-related Virus Mutations, Integration sites and Cis-effects)
The ViMIC is a resource of human disease-related virus information. It mainly covers three special features, including virus mutation sites (VMs), viral integration sites (VISs) and target genes. The database allows users to quickly query the information regarding curated VMs, VIS-Cistrome intersections, statistics of virus sequences data, VIS-VM associations and the correlation between immune cell infiltration levels and the expression of genes which are affected by the integration event or virus gene/region/protein in viral infectious diseases.
Help Video
We recommend using google chrome to watch this video.
Help Documentation Contents
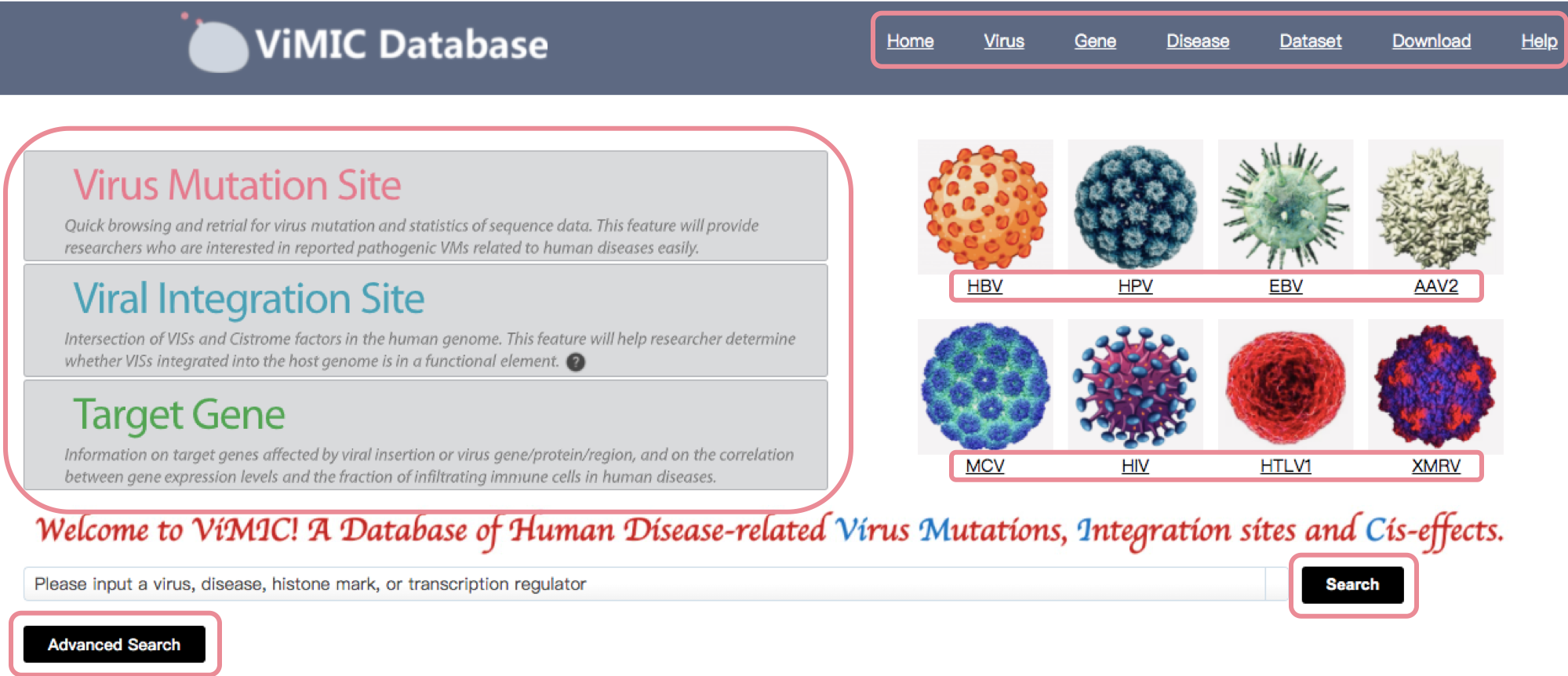
Homepage
The homepage of ViMIC mainly gives a navigation of our database, including listing the modules of ViMIC on menu bar and upper left panel, providing three main features to help users to browse, search, analyze, visualize and download data. Main modules on upper left consist of virus mutation site, viral integration site and target gene. Other modules consist of Virus (virus information summary), Gene (target gene summary), Disease (virus infected disease summary), Dataset (virus related clinical dataset summary), Download (ViMIC data download) and Help (ViMIC help document).
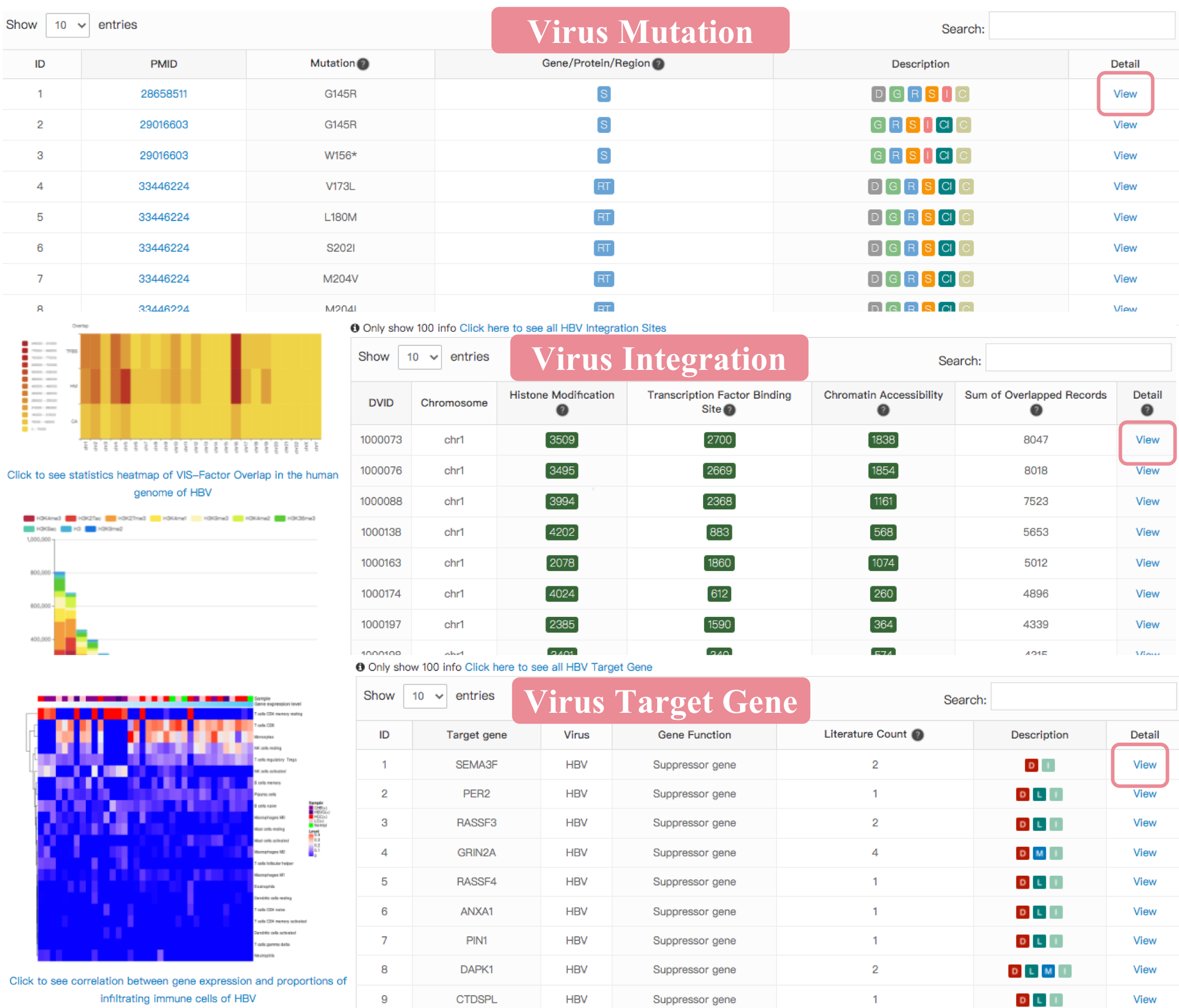
Virus Mutation
Overview of Mutation
The “Overview of Mutation” page provides the curated mutation information for each virus. The table shows ‘Mutation Entries’ and virus associated ‘Disease Name’ in each virus which are curated by ViMIC. Click ‘Virus Name’ to show the information of mutations, VIS-Cistrome factor overlapped records and target genes of selected virus. Click ‘Disease Name’ to display the summary of literature related to selected virus associated disease. Click ‘Mutation Entries’ to show the detailed mutation information of selected virus.
Virus Navigation and Mutation Statistics

Virus Mutation Site
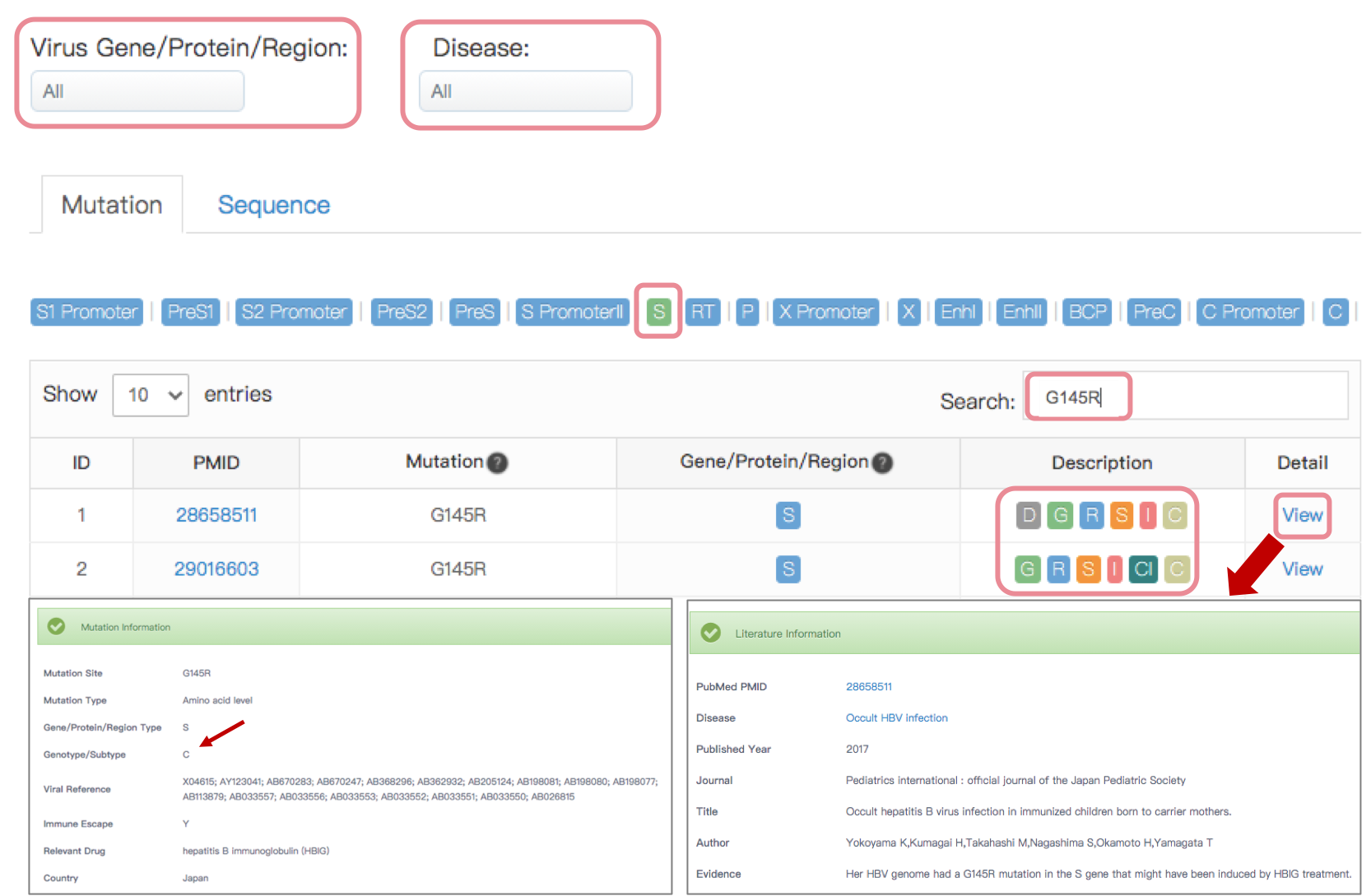
The VM annotation module provides researchers who are interested in VMs with annotations of reported pathogenic mutations in virus related to human diseases easily. The curated mutation information includes mutation site, mutation level, virus gene/protein/region, virus-associated disease, genotype/subtype, literature evidence, etc. Two drop-down menu items above the table for users to choose different virus gene/protein/region and disease browsing options respectively. The table below shows curated mutation sites of selected virus.
We designed a description column to indicate more detailed information for each mutation site using the following tags:
• D: Drugs mentioned in the relevant literature;
• G: Genotypes/subtypes mentioned in the relevant literature;
• R: Reference genomes mentioned in the relevant literature;
• S: Viral sequences mentioned in the relevant literature;
• I: This mutation is related to the immune escape;
• Cl: Clinical information is available in the relevant literature;
• C: Geographical distribution of the source mentioned in the relevant literature.
When selecting the virus gene of interest, users can quickly search the keyword of interest in the search box on the top-right of the table. Click the "View" button to show the detail information of this mutation.
Statistics of Virus Sequences Data
The Viral Sequence module provides the geographical distribution of the source, viral gene, region and genotype of virus sequences. Users can browse the statistical data and download the annotation information to quickly obtain the sequence GeneBank IDs of interest.

Viral Integration
Overview of Virus Integration Site (VIS)-Cistrome Factor Feature
This feature provides information of the overlapped records between VIS and Cistrome factors as well as Cistrome factor-related samples, the associated factor and overlap amount could indicate that the virus integration site is a potentially functional DNA element. The cistrome refers to "the set of cis-acting targets of a trans-acting factor on a genome-wide scale." The table below provide virus navigation and VIS-cistrome data statistics which summaries about VIS we collected as well as VIS-Cistrome factor overlapped records in each virus. Click virus name to view virus overview of virus mutation, integration and target gene. Click VIS count number (green button) to view VIS-Cistrome factor data. Click Disease Information to display the summary of this disease and disease related literature. The user can click "Search VIS-Cistrome Overlap" to Search Overlapped Records of Interest page with saerch engine for saerching overlapped records of interest according to disease, biological resource (cell line, cell type, tissue type) and other selections. Click "Explore VIS batch (list)" to search VIS batch or upload VIS list to ViMIC.

Search Overlapped Records of Interest
This search module can help users search and filter the overlapped records of interest according to various keywords (e.g., virus name, chromosome, location, disease, Cistrome biological source or Cistrome type). Specifically, the user can combining keywords of related disease and biological resource to filter out overlpped records of interest. ViMIC will then return a factor grouped table containing overlapped records which meet all selected options that the user is interested in.
For example, if the user want to filter out overlap data that transcription factor binding site of hepatocellular carcinoma cell in conjunctions with Hepatitis B virus integrations with hepatocellular carcinoma on chromosome 1:
1. Select HBV, chromosome 1, hepatocellular carcinoma, transcription factor binding site on four combo boxes in turn, then type or seclect hepatocellular carcinoma (cell type) on last text box and click submit.
2. A search result page will automatically opened in new tab, the table shows a factor grouped table containing overlapped records which meet all options selected in step 1.
3. The user can click green button with number to view GSMID (or ENCODE) and biological source detail.

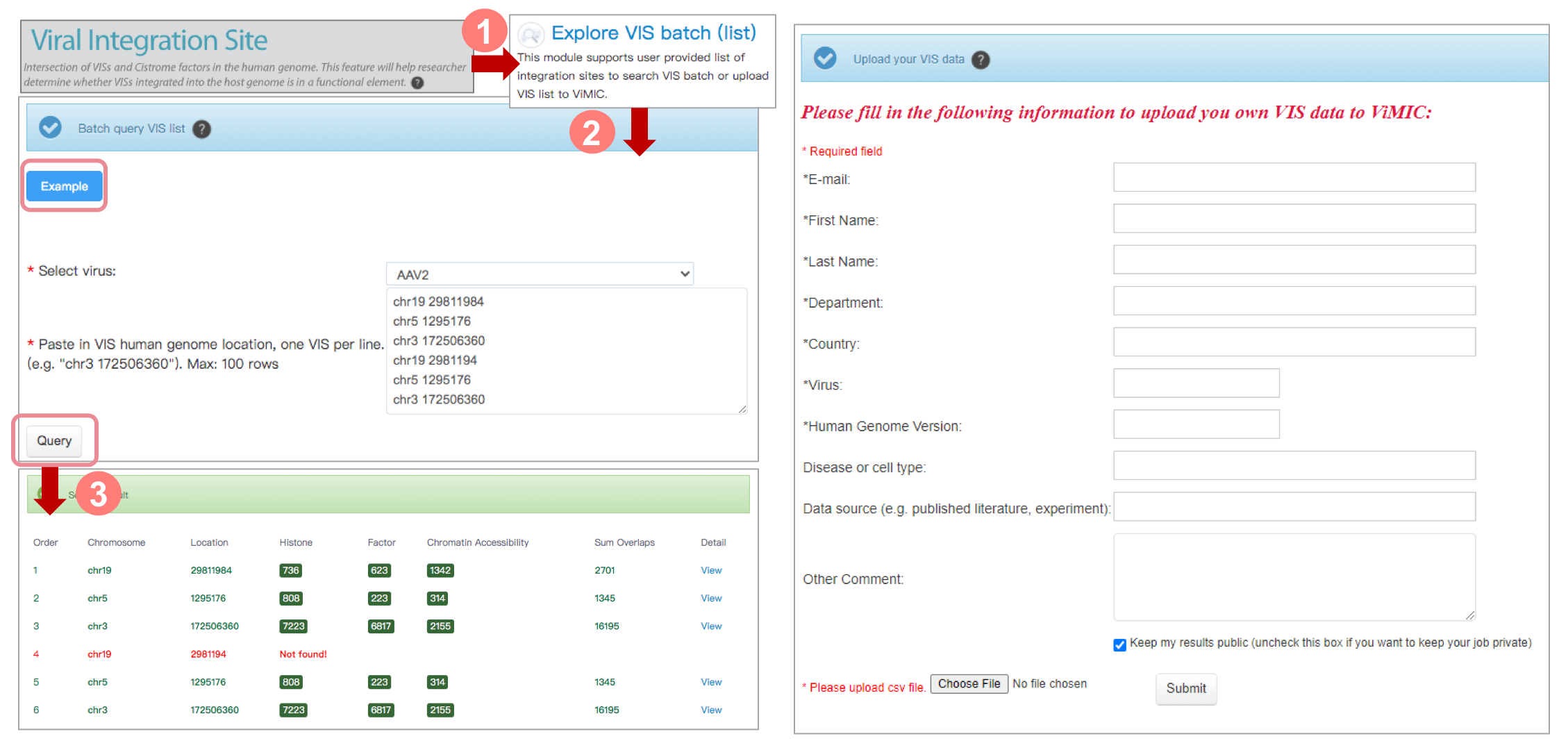
Explore VIS batch (list)
This module supports user provided list of integration sites to search VIS batch or upload VIS list to ViMIC. The result will be notified by email once the job is completed if an email address has been provided.
VIS-Cistrome Factor Overlap in each human chromosome
• Overall Heatmap
Here, take HBV as an example. This heatmap is an overall statistics for HBV VIS-Cistrome factor overlapped records by chromosome. An overlapped record means a match between VIS and 1 of 3 Cistrome factors (histone modification, transcription factor binding site and chromatin accessibility), more specificly, this match includes at least 1 overlap between this VIS and 1 Cistrome factor-related sample from 1 of 3 Cistrome factor data. The darker the red, the greater the overlapped records number. The user can view specific number by moving mouse pointer.
• Browse Data by Chromosome or Factor
The user can browse HBV VIS-Cistrome factor overlapped records data by specific chromosome or statistics histogram for specific Cistrome factor when click the chromosome or Cistrome factor the user is interested in.
• VIS locations with VIS-cistrome overlap information and VIS details for each chromosome
This table shows VIS locations with VIS-cistrome overlap information for each chromosome. When selecting the chromosome of interest, users can browse VIS information which indicates the numbers of VIS and three cistrome factors’ overlapped records. Click ‘View’ button to show the detailed information of selected VIS.
By clicking “Show VIS with two points” button, ViMIC will return VIS-cistrome table of VIS with two points. Columns with “(S)” are overlaps calculated by single start site and columns with “(B)” are overlaps calculated by both two sites. Column “Case/Length” shows the case classification of VISs with two sites based on the original literature and length between two sites.
1. Case 1: The deletion of human fragments during the integration event.
2. Case 2: Due to the experimental method/design, two viral insertion positions were provided in the literature.

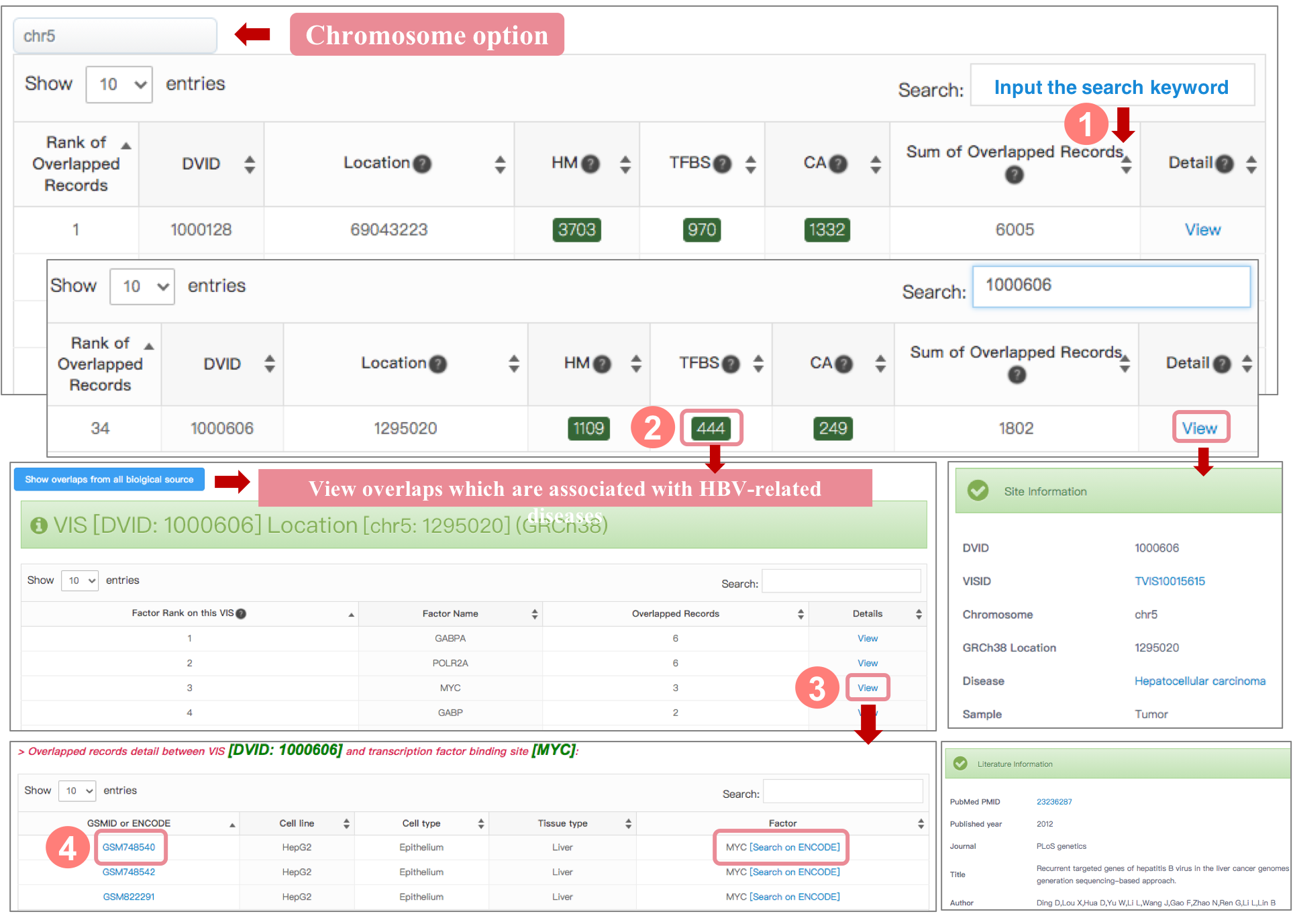
VIS-Cistrome Factor Overlapped Record Details
This table at top of page is factor ranked by overlap number. The overlap number refers to the overlapping count between VIS selected and one sepecific cistrome factor. The user can click biological source filter with blue button to keep the overlaps which are associated with virus-related diseases based on cistrome sample's biological source.
An overlapped record means a match between VIS and 1 of 3 Cistrome factor (histone modification, transcription factor binding site and chromatin accessibility), more specifically, this match includes at least 1 overlap between this VIS and 1 Cistrome factor-related sample from 1 of 3 Cistrome factor data. So one VIS may have several overlaps with multiple Cistrome factor-related samples, the associated factor and overlap amount of samples could indicate that the virus integration site is a potentially functional DNA element.
The interface details are as follows:
1. We take the entry of “1000606” on chromosome 5 in HBV as an example. After users click the “chr5” and search the entry of “1000606” in HBV VIS-Cistrome Factor Overlap in each human chromosome page, ViMIC shows the overlaps result of three cistrome factors. The user can click green button to view VIS-Cistrome Factor Overlapped Record Details related to selected VIS, in addition, the user can inspect some other VIS detail information such as disease and literature by clicking "View" button at Detail column.
2. By clicking the green button with number at “TF” (Transcription factor binding site) column, ViMIC shows the ranking of all transcriptional regulators that have overlaps with this VIS.
3. In this case exploration, users can view cistrome samples by clicking the ‘View’ button of the transcription factor MYC, and then users can view the detail information of samples (GSMID, cell line, cell type, tissue type and factor name)
4. Additionally, sample ID and factor name are crosslinked to the related resources, including GEO and ENCODE databases.
Virus Integration Site and Cistrome Overlapped in each chromosome on human genome
The histogram shows the overlap count of three factors (histone modification, transcription factor binding site, and chromatin accessibility) and virus intergration fragments on the human chromosomes. The y-axis represents the number of overlap, and the X-axis represents the integrated fragments. The table shows the Cistrome factor count rank of VIS on the chromosome user selected. The user can click green button to view details and view VIS detail information by clicking "View" button at Detail column.

Integration Site Overlapped Cistrome Factor Summary
This page summarizes the count of VIS-Cistrome factor overlapped records and the number of VIS which have at least one overlap with each Cistrome factor on the human chromosomes. It also provides top ranked Cistrome factors ordered by the overlap number in each chromosome using stacked barplot.
• Top 10 Cistrome factor ranked by the Overlap Sum
Histogram of factors by chromosome. The statistic factors are top 10 amount factors on all HBV VIS-Cistrome factor overlapped records data. Strategy: Firstly, rank the overlapped records by Cistrome factor, then only visualize the top 10 Cistrome factor statistics to histogram by Chromosome.
• Top 5 Cistrome factor ranked by the Overlap in each chromosome
Histogram of factors by chromosome. The statistic factors on are the sum of the top 5 amount factors on each Chromosome. Strategy: Firstly, rank the overlapped records by Cistrome factor for each Chromosome, then only visualize the sum of top 5 Cistrome factor statistics of each Chromosome to histogram by Chromosome.
• Factor Statistics
The table summarizes the number of VIS which have at least one overlap with each Cistrome factor on the human chromosomes.

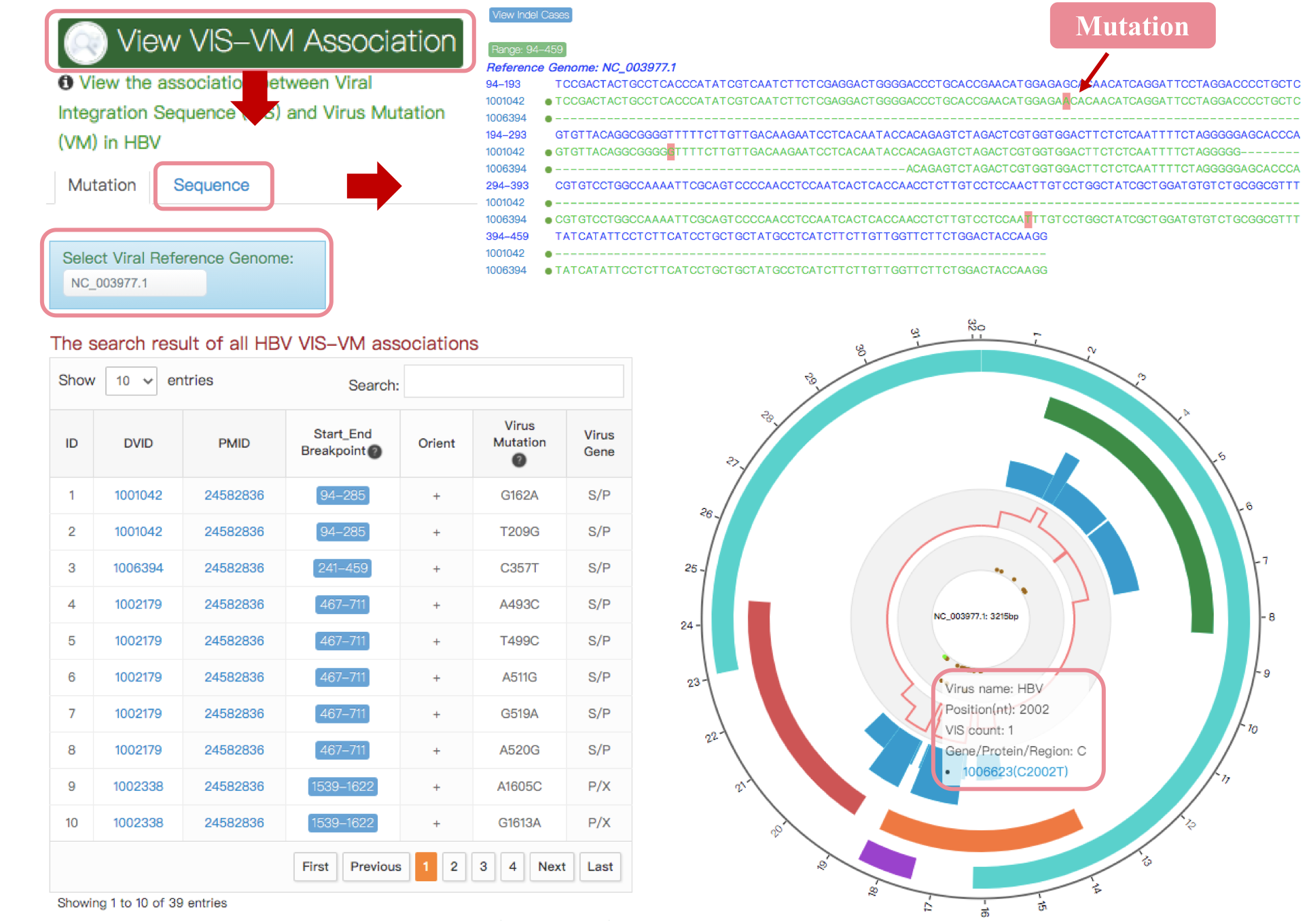
VIS-VM Association
In the VIS-cistrome homepage of selected virus, users can click “View VIS-VM Association” to visualize mutations observed in VIS fragments. VISs of 3 viruses includes HBV, HPV and AAV2, with precise breakpoint coordinates and the reference genome in the integration literature, were used to map and visualize whether mutation occurred in VISs. Users can view the distribution of mutations harbored in VISs compared with the viral reference genome (e.g., NC_003977.1) sequence and downlaod all viral VIS-VM associations results. Users can click DVID to query the detailed information of the VIS. A virus genome plot on the right visualizes the distribution of VISs based on each viral reference genome. Move your mouse over the plot to view the region and VIS information. By clicking ‘Sequence’ button, users can view the sequence mapping of identify mutations harbored in each VIS.
Return to Top
Target Gene
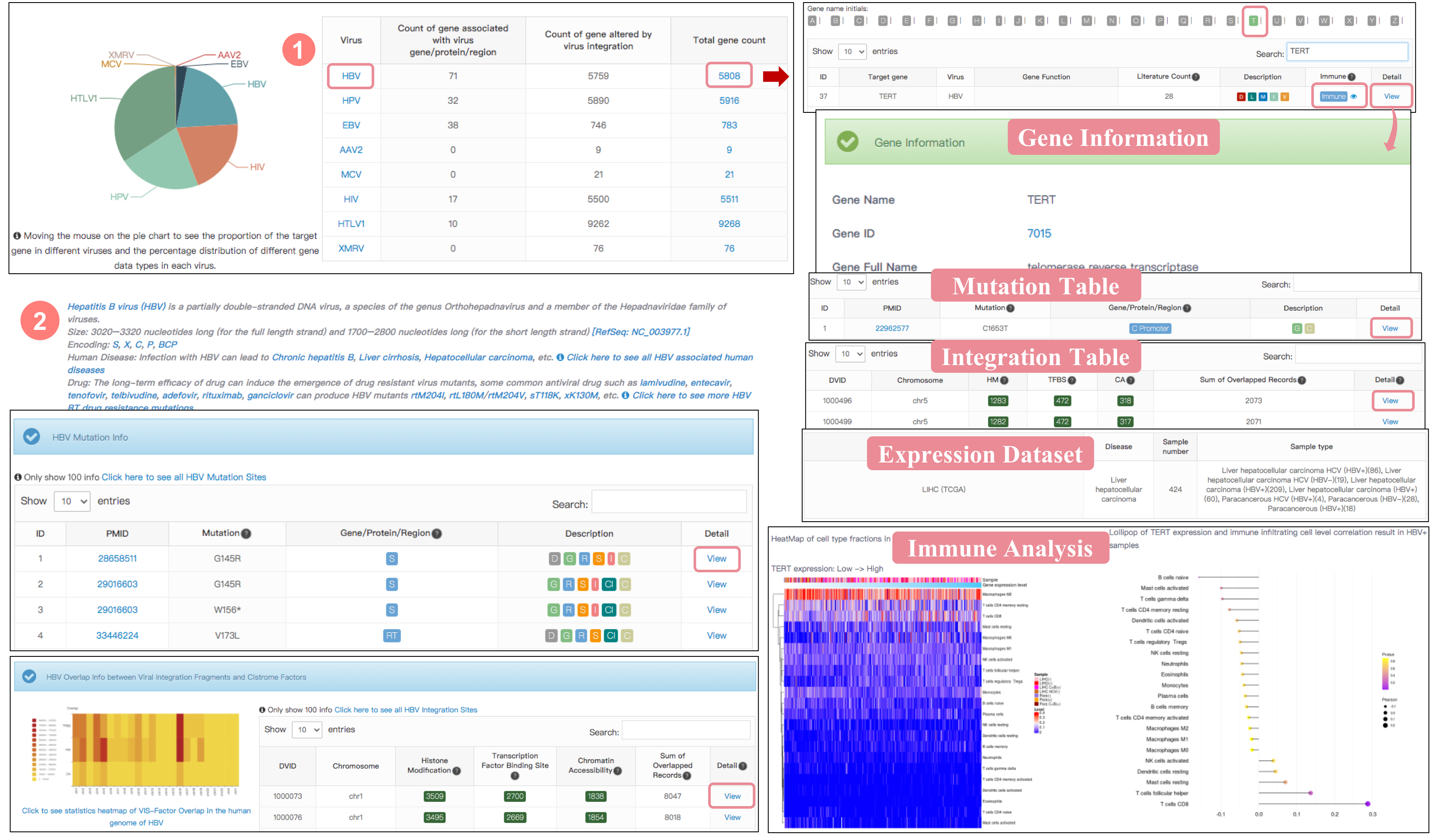
Overview of Virus Target Gene
The “Target gene” feature provides information for the target gene affected by viral insertion or virus gene/protein regulation. The detail information also presents the correlation between gene expression levels and proportions of 22 types of infiltrating immune cells in human disease. A pie chart shows the number of the target gene in each virus and the percentage distribution of different gene data types. Click virus name (blue font) to see this virus introduction, virus mutation, integration-cistrome factor overlap and target gene. The table contains the sum of genes associated with virus gene/protein/region and virus integration in each virus, and provides the detail information of gene type, the relevant literature count, protein location, immune-related analysis, etc.
Target gene Detail Information
Gene information mainly shows gene name, gene id, gene type, resources' ID, etc, tow table (the integration table and gene expression dataset table) and immune analysis plot. The integration table: if previous studies reported that target gene was altered by virus integration events, the overlap between VISs in this literature and Cistrome factors was listed in the table. The gene expression dataset table lists the information of gene expression file from GEO or TCGA which contain the targetgene expression data, including data ID, disease, sample number and sample type.
Immune analysis plot mainly has two parts, Left: heatmap of target gene expression and the level of various types of immune cells (cell type fractions) in all samples based on CIBERSORT method. Right: correlation of target gene expression and the level of various types of immune cells in virus positive samples (Pearson Correlation Coefficient).
Return to Top
Virus Overview
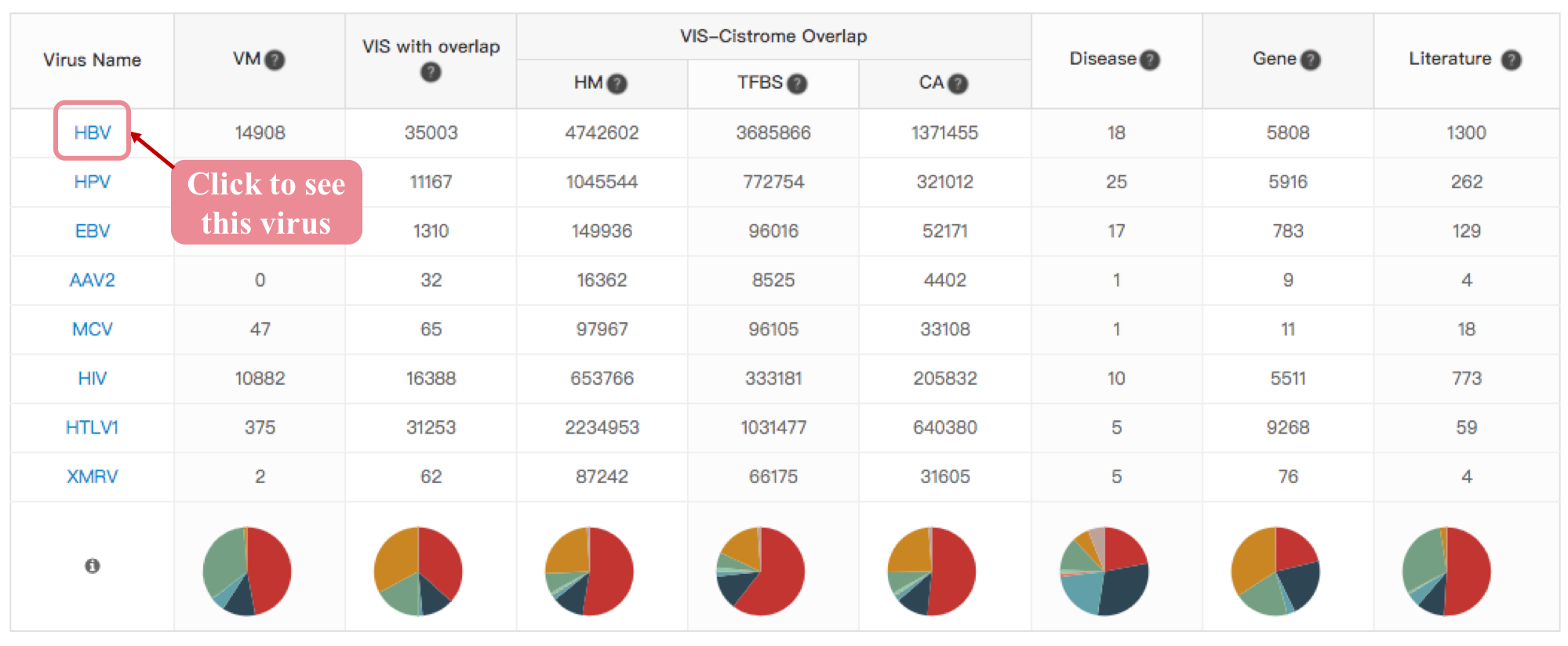
Virus Info Summary
The Virus module provides an interface of each virus with a summary and links to three ViMIC features to help users easy to query research questions on VMs, VISs-Factors overlap or target genes, the table below shows the detailed count of each statistical type, click the virus name in table to see the overview of HBV mutation, integration-cistrome and target gene.
Overview of Virus Mutation, Integration-Cistrome Factor Overlap and Target gene

This page gives an overview of Virus Mutation, Integration-Cistrome Factor Overlap and Target gene. Mutation includes partial data of mutation sites. Integration includes heatmap of VIS-Cistrome overlap count in the human genome, histogram of cistrome factors by chromosome, distribution of VIS-Cistrome overlapped in chromosome and table of VIS information. Target gene includes correlation diagrams of gene expression and proportions of infiltrating immune cells as well as target gene information table. Click the blue font on the page to view detail information.
Return to Top
Virus Associated Disease
Virus Infected Disease Summary
The Disease module provides the curated information in each type of virus related disease, including mutation, integration-cistrome and literature. Mutation count means the total number of virus mutation in virus associated diseases. Integration count means the total number of virus integration in virus associated diseases. Literature count means the total number of literature related to virus associated diseases.
Overview of Disease
The table shows literature information of this disease.
Two related tag:
• Mutation Tag: literature about this virus mutation and click the eye icon to see detail mutation information in literature.
• VIS-cistrome Tag: literature associated this virus integration and click the eye icon to see detail information of 3 cistrome factors (histone modification, transcription factor binding site and chromatin accessibility).

Return to Top
Virus Dataset
Virus Dataset Summary
This page shows statistical charts and tables about descriptive clinical information for virus mutation and integration datasets. The user can click "View" to inspect detailed clinical information and some links for the sample.

Return to Top
Search
ViMIC mainly provides two search ways: quick search and advanced search.
Quick Search
Input the virus name, disease, histone mark, transcription regulator or gene/protein/region directly in the search box at homepage, poping out a window about results of searched words.
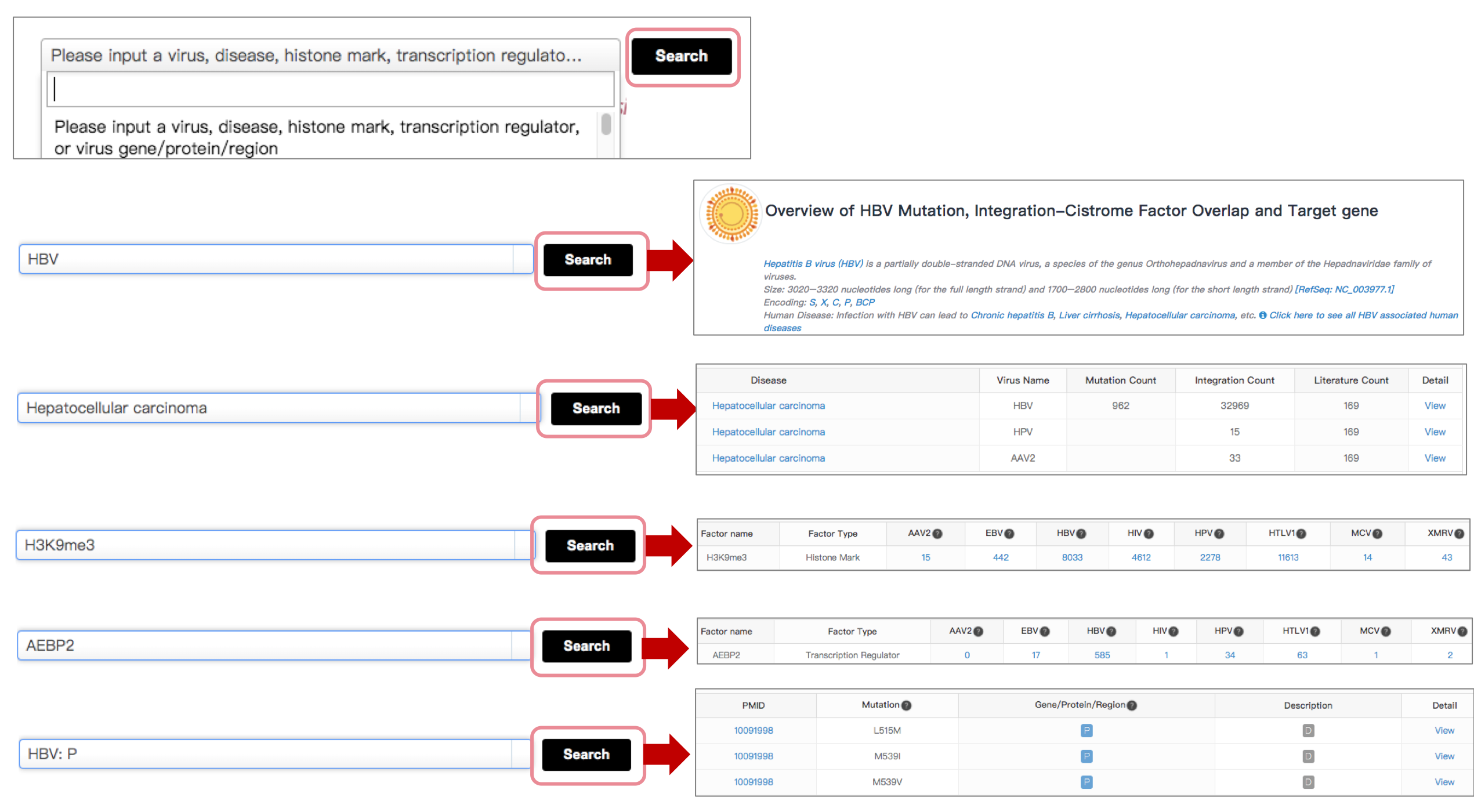
Advanced Search
Advanced search is available for sophisticated searches by combining the various keywords (Virus, Mutation Site, Virus Gene/Protein/Region, Disease, Chromosome, Fragment) when user click Advanced Search on the left side of the homepage. Advanced search option for sophisticated searches by combining the various keywords (e.g., virus name, chromosome, virus gene/protein/region or target gene).

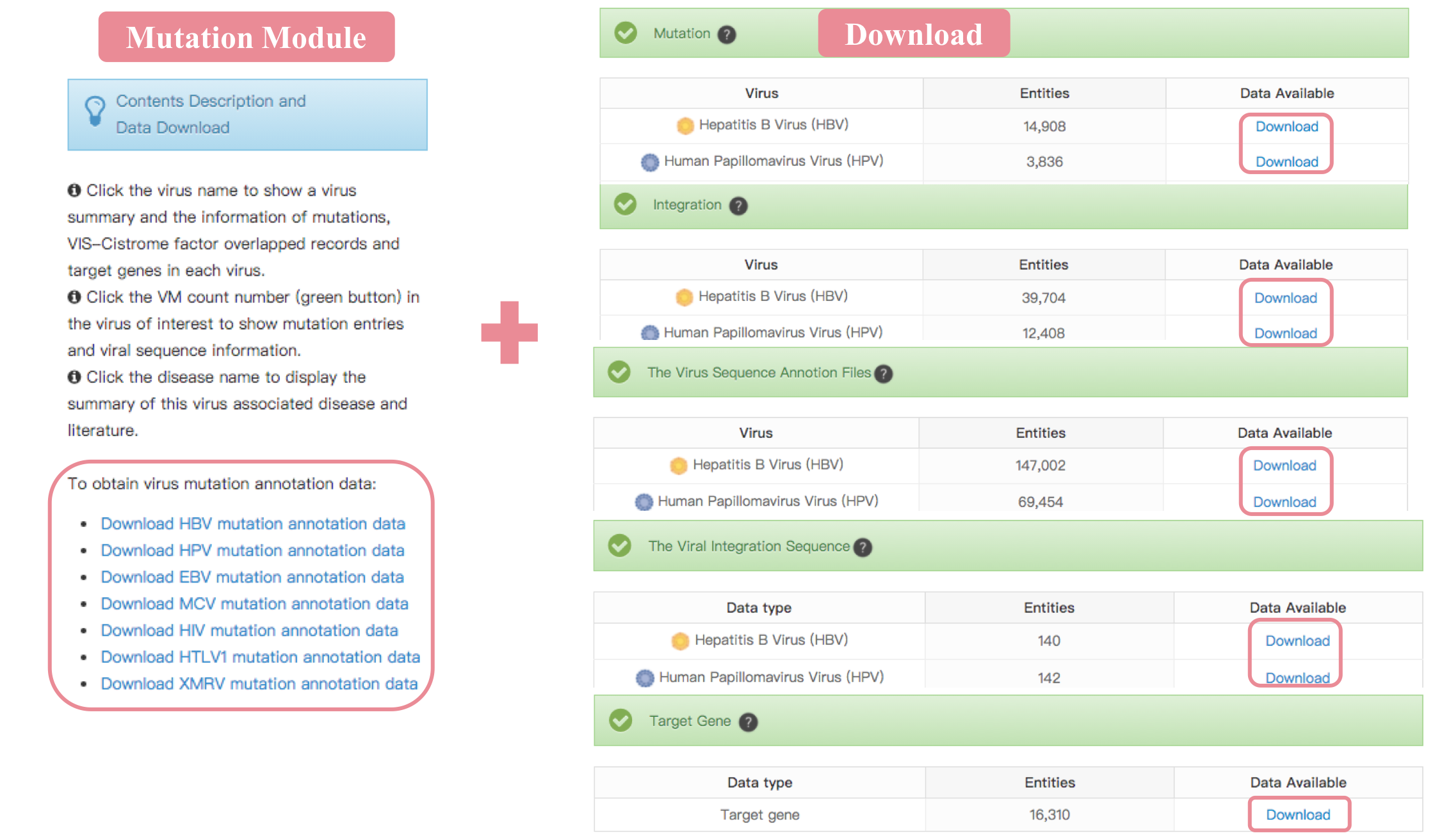
Download
The ViMIC allows the user to download the various virus data files including mutation, integration, sequences and target gene. Meanwhile, users can directly download the related data files in the mutation, integration, target gene module pages or data download page.
Contact us
Emails
Xiaoyan Zhang: xyzhang@tongji.edu.cnAddress
Tongji University, No.1239, Siping Road, Shanghai, P.R. ChinaReturn to Top