|
Gene Information
|
Gene Name
|
FGF2 |
|
Gene ID
|
2247
|
|
Gene Full Name
|
fibroblast growth factor 2 |
|
Gene Alias
|
BFGF; FGFB; FGF-2; HBGF-2 |
|
Transcripts
|
ENSG00000138685
|
|
Virus
|
HTLV1 |
|
Gene Type
|
protein coding |
|
HPA Location Info
|
Nuclear bodies;Nucleoplasm;Predicted to be secreted; 
|
|
HPA Protein Class Info
|
Cancer-related genes, Candidate cardiovascular disease genes, FDA approved drug targets, Predicted intracellular proteins, Predicted secreted proteins, RAS pathway related proteins, Transporters |
|
Uniport_ID
|
P09038
|
|
HGNC ID
|
HGNC:3676
|
|
VEGA ID
|
OTTHUMG00000039506
|
|
OMIM ID
|
134920 |
|
String ID
|
9606.ENSP00000264498
|
|
Drugbank ID
|
BE0001015
|
|
PharmGKB ID
|
PA28115
|
Target gene [FGF2] related to VISs 
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 6000302 |
chr4 |
1142 |
622 |
487 |
2251 |
View |
Relationship between gene expression and immune infiltrating cell level
| Data ID |
Disease |
Sample number |
Sample type |
|
GSE55851 (GEO) |
Adult T-cell leukemia |
21 |
Normal (3), Asymptomatic carriers (HTLV1+)(6), Adult T-cell Leukemia-lymphoma (HTLV1+)(12) |
|
GSE29312 (GEO) |
HTLV-1 associated myelopathy |
39 |
Normal (9), Asymptomatic carriers (HTLV1+)(20), HTLV-1-associated myelopathy (HTLV1+)(10) |
|
GSE29332 (GEO) |
HTLV-1 associated myelopathy |
35 |
Normal (8), Asymptomatic carriers (HTLV1+)(17), HTLV-1-associated myelopathy (HTLV1+)(10) |
> Dataset: GSE55851
|
|
HeatMap of cell type fractions in all samples
|
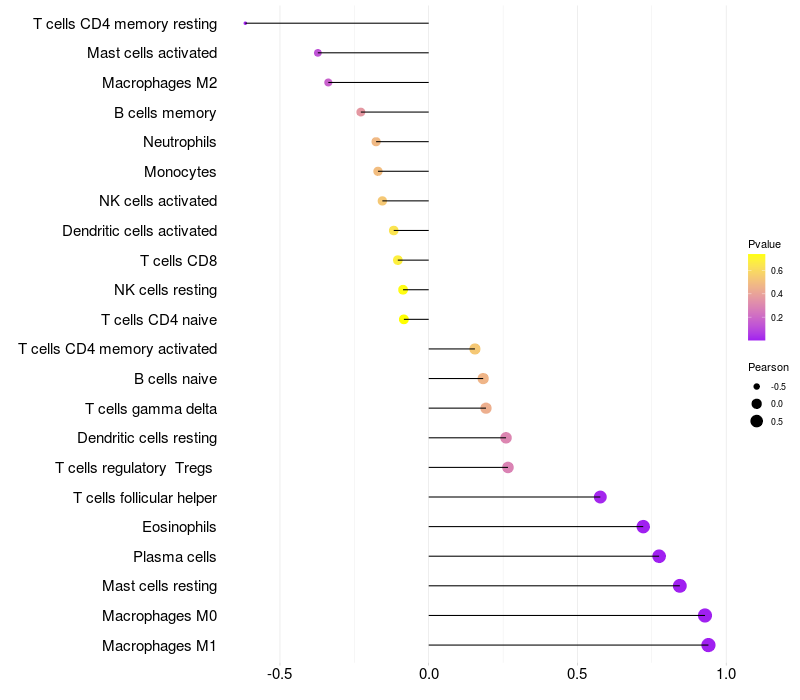
Lollipop of FGF2 expression and immune infiltrating cell level correlation result in HTLV1+ samples
|
FGF2 expression: Low -> High
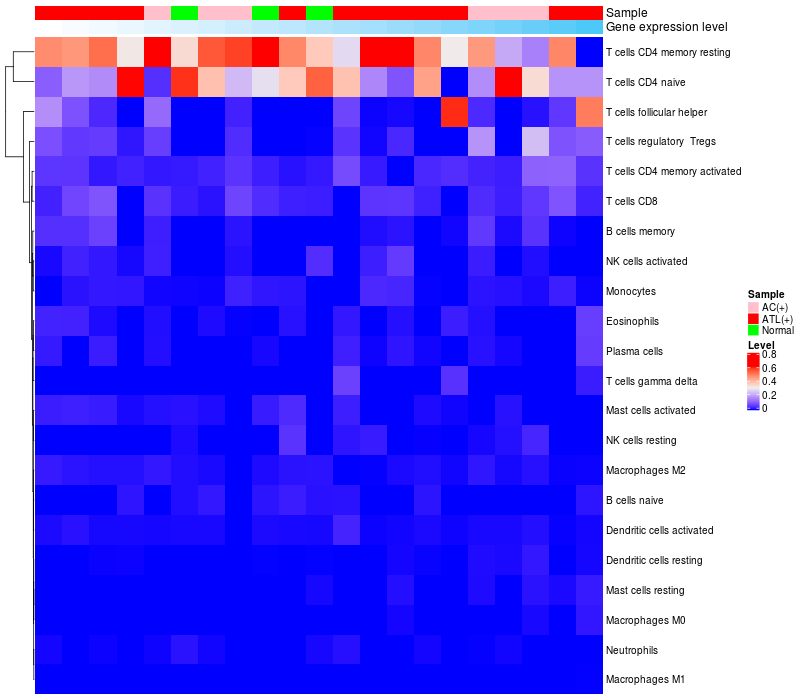
|
|
|
> Dataset: GSE29312
|
|
HeatMap of cell type fractions in all samples
|
Lollipop of FGF2 expression and immune infiltrating cell level correlation result in HTLV1+ samples
|
FGF2 expression: Low -> High
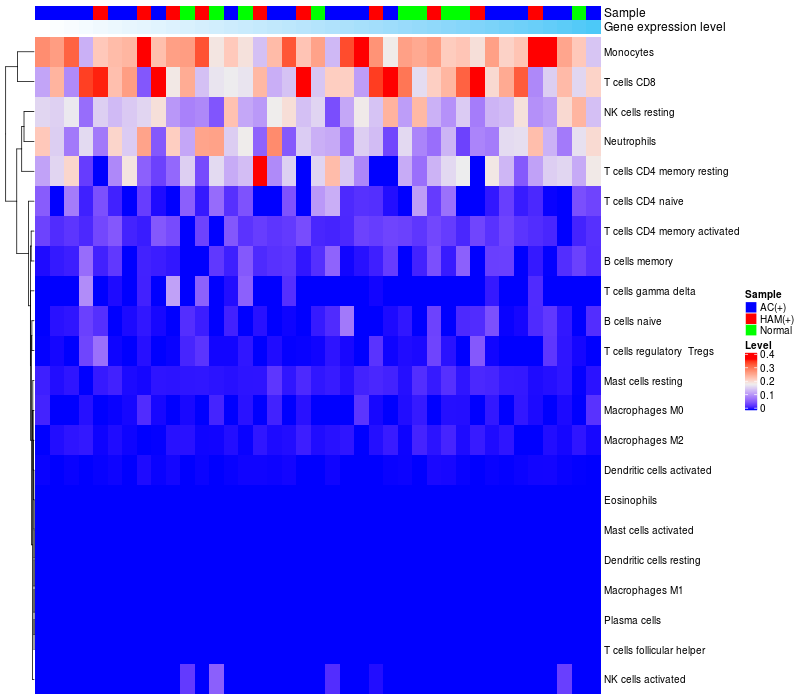
|
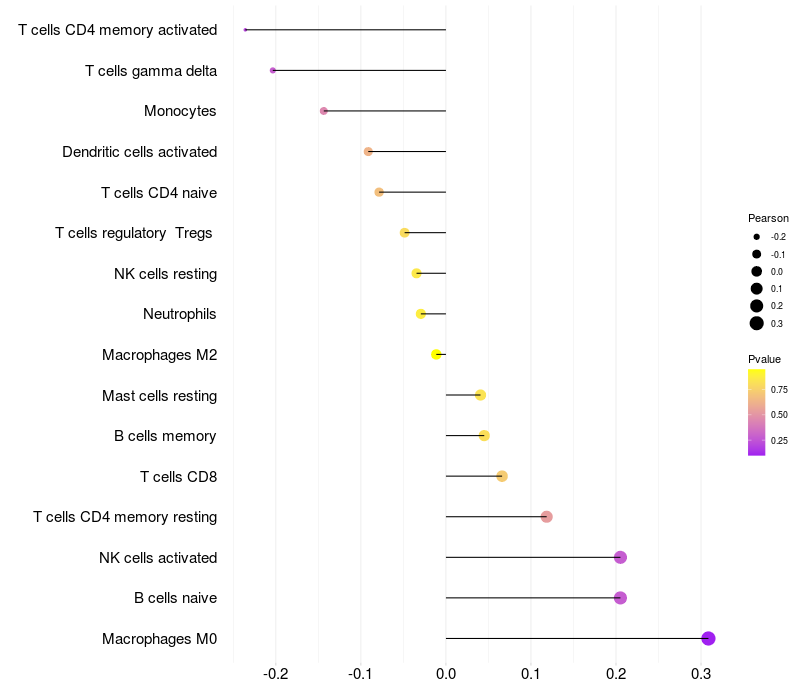
|
|
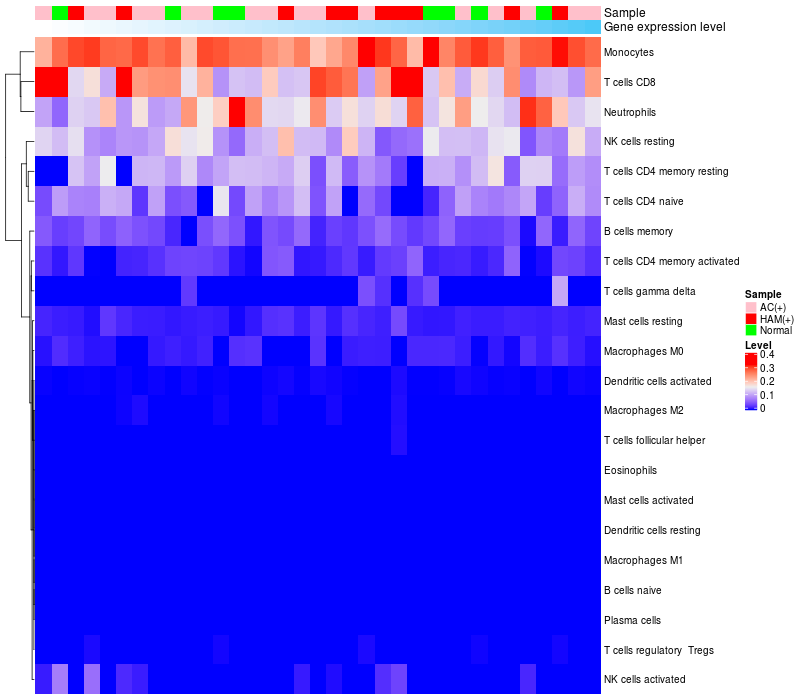
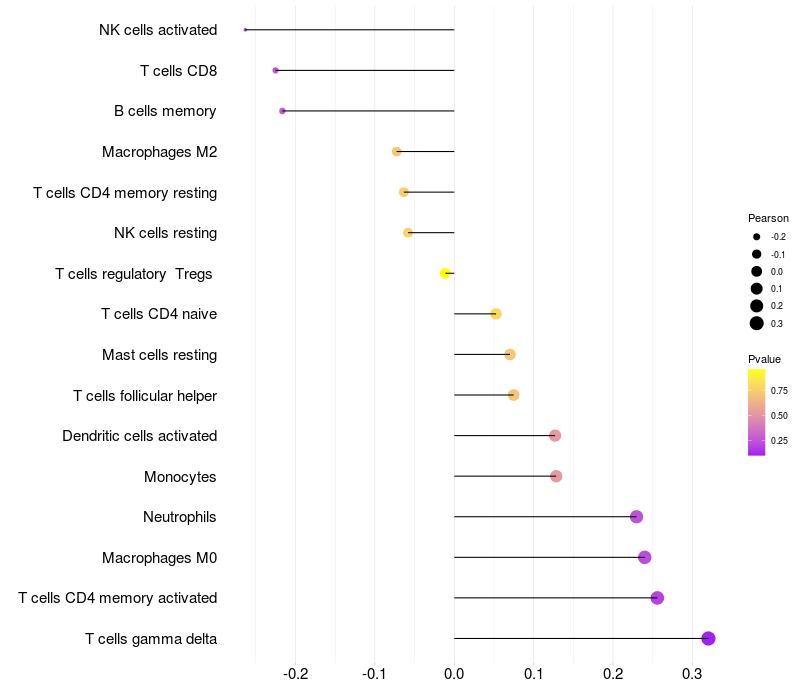
> Dataset: GSE29332
|
|
HeatMap of cell type fractions in all samples
|
Lollipop of FGF2 expression and immune infiltrating cell level correlation result in HTLV1+ samples
|
FGF2 expression: Low -> High
|
|
|
|
|
 HTLV1 Target gene Detail Information
HTLV1 Target gene Detail Information