|
Gene Information
|
Gene Name
|
CALCRL |
|
Gene ID
|
10203
|
|
Gene Full Name
|
calcitonin receptor like receptor |
|
Gene Alias
|
CRLR; CGRPR |
|
Transcripts
|
ENSG00000064989
|
|
Virus
|
HBV |
|
Gene Type
|
protein coding |
|
HPA Location Info
|
Plasma membrane; 
|
|
Membrane Info
|
Predicted membrane proteins |
|
HPA Protein Class Info
|
G-protein coupled receptors, Plasma proteins, Predicted intracellular proteins, Predicted membrane proteins, Transporters |
|
Uniport_ID
|
Q16602
|
|
HGNC ID
|
HGNC:16709
|
|
VEGA ID
|
OTTHUMG00000132636
|
|
OMIM ID
|
114190 |
|
String ID
|
9606.ENSP00000386972
|
|
Drugbank ID
|
BE0008934
|
|
PharmGKB ID
|
PA26033
|
Target gene [CALCRL] related to VISs 
| DVID |
Chromosome |
HM |
TFBS |
CA |
Sum of Overlapped Records |
Detail |
| 1009873 |
chr2 |
1 |
8 |
5 |
14 |
View |
| 1014750 |
chr2 |
3 |
|
|
3 |
View |
Relationship between gene expression and immune infiltrating cell level
| Data ID |
Disease |
Sample number |
Sample type |
|
LIHC (TCGA) |
Liver hepatocellular carcinoma |
424 |
Liver hepatocellular carcinoma HCV (HBV+)(86), Liver hepatocellular carcinoma HCV (HBV-)(19), Liver hepatocellular carcinoma (HBV+)(209), Liver hepatocellular carcinoma (HBV+)(60), Paracancerous HCV (HBV+)(4), Paracancerous (HBV-)(28), Paracancerous (HBV+)(18) |
> Dataset: LIHC
|
|
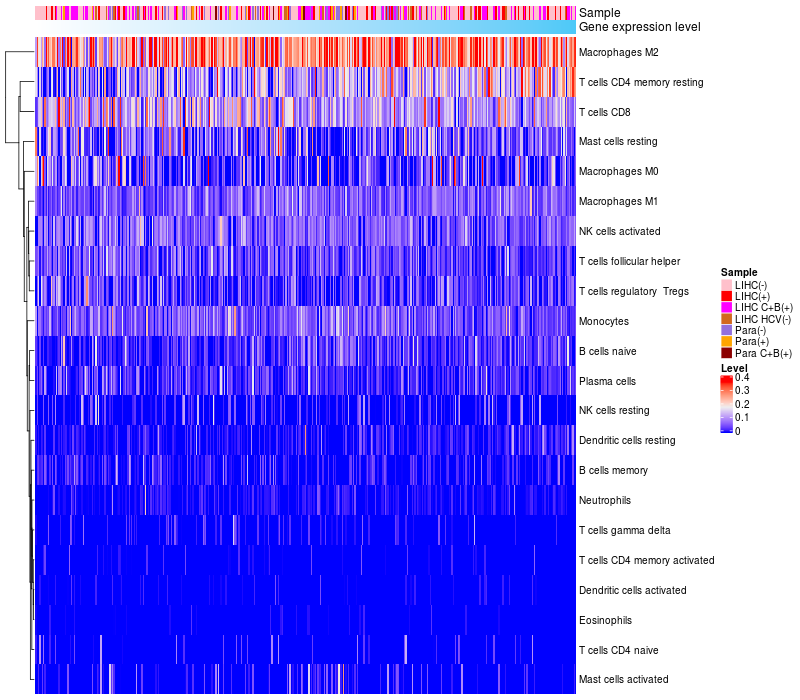
HeatMap of cell type fractions in all samples
|
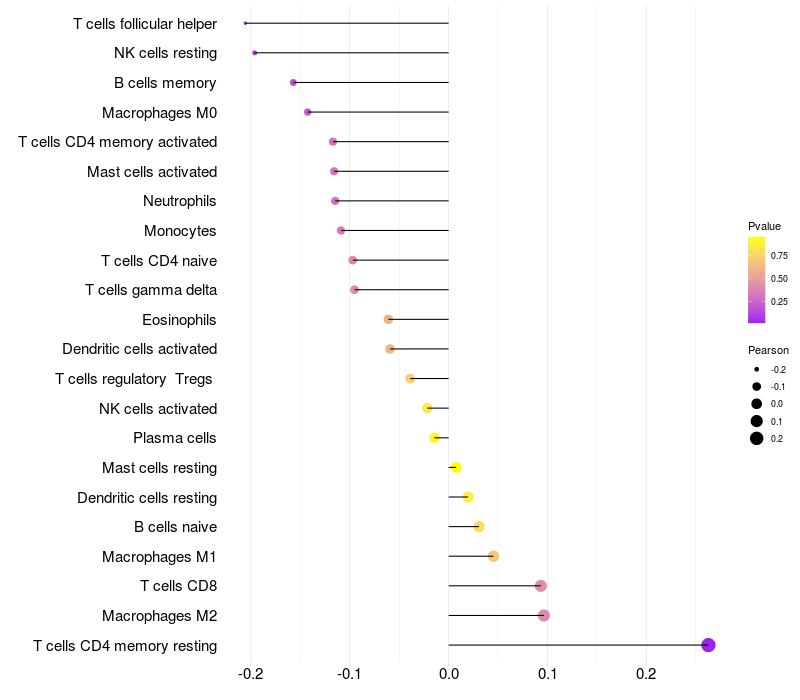
Lollipop of CALCRL expression and immune infiltrating cell level correlation result in HBV+ samples
|
CALCRL expression: Low -> High
|
|
|
|
|
 HBV Target gene Detail Information
HBV Target gene Detail Information