Documentation
Our paper [N/A]Introduction
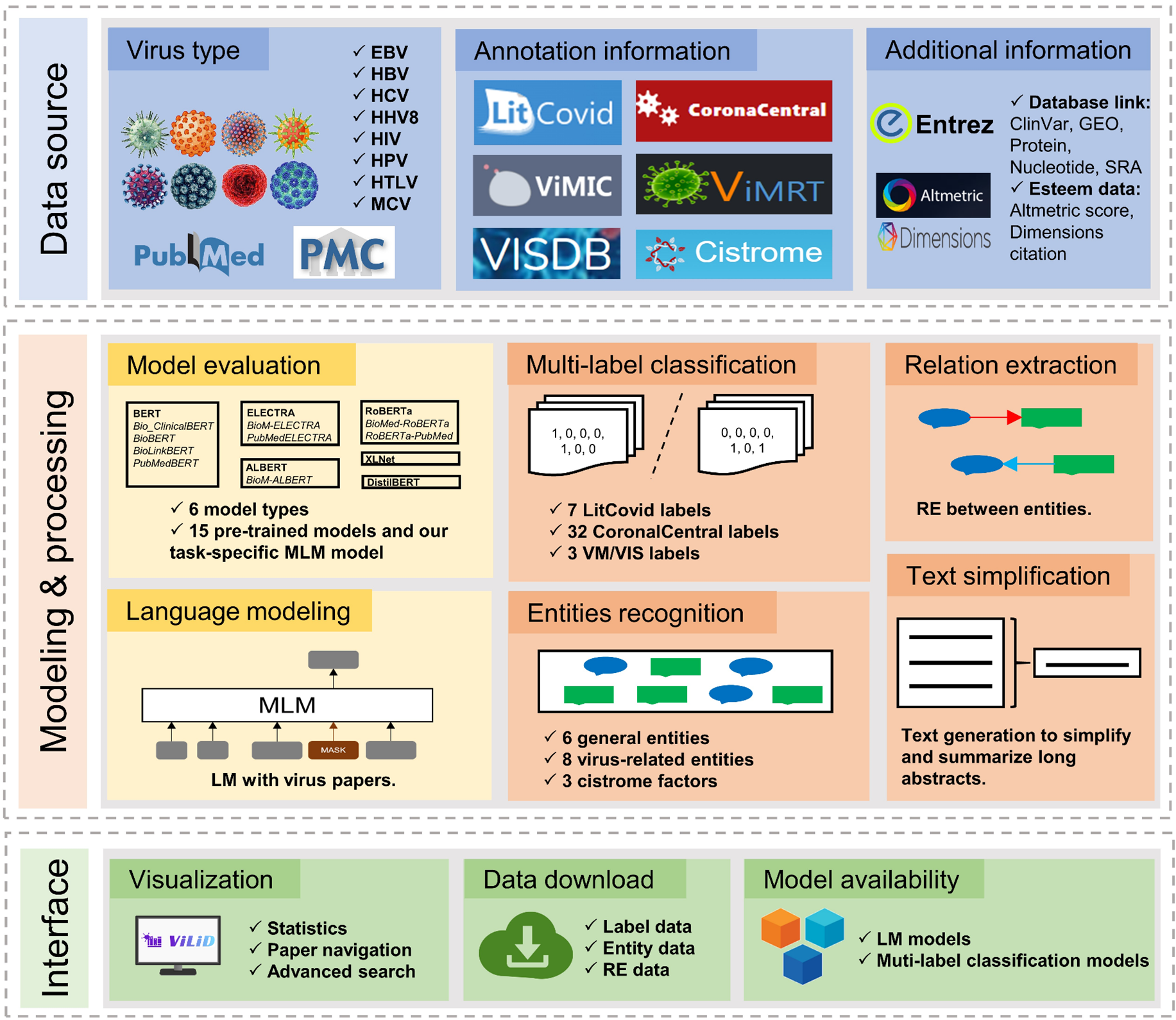
Study overview
First, at the data source level, the literature and its label data of several databases, as well as manual annotations were used to build the deep learning dataset, while we obtained virus literature data from PubMed as a corpus, and also integrated data from five genomic databases using Entrez and included Altmetric score data as supplementary data for the literature. Second, in the model implementation part, we evaluated the performance of 14 pre-trained language models by fine-tuning them on the multi-label classification task of virus topics and used the unlabeled raw text of the virus corpus to pre-train the best-performing pre-trained model with masked language modeling to further improve the performance. We fine-tuned the virus-specific pre-trained model on three multi-label classification tasks, and predicted the probability of each label for the large-scale literature of 8 severe disease-related viruses, providing a basis for label classification in the literature database. In addition to the semantic topic label prediction of the literature, we also identified general entities (such as genes, diseases, drugs, etc.), virus-related entities (such as virus transmission types, detection types, vaccine types, etc.), and cistrome factors through entity recognition, which are used in predicting on inter-entity relations. Moreover, we used text generation models for summarizing key points of long abstracts. Third, we have built a publicly available web interface with all curated data, allowing users to quickly browse and navigate the literature of interest, and provide downloads of all data and models.
About ViLiD
In ViLiD, firstly, users can quickly access the literature of interest in 7 major topics (e.g., mechanism, treatment), as well as 32 detailed topics (e.g., drug targets, imaging, vaccines) that can help users access more specific literature, moreover, users can access literature related to VM and VIS. In addition, users can access information of 5 types of genome data (ClinVar, GEO, Protein, Nucleotide, SRA) related to the literature. Secondly, ViLiD provides entity recognition of the literature (including 6 general entities, 8 virus-specific entities, and 3 cistrome factors) and provides predictions of relation extraction between entities. Besides, ViLiD provides text generation and rewriting for long abstracts to summarize the key points, which helps users to quickly comprehend the focus of the paper. Thirdly, users can use advanced search to qualify all labels, entities, datasets, etc. to quickly find the literature that meets the criteria of interest. More importantly, the pre-training model, fine-tuning model and data of database developed by ViLiD are publicly available in the download module, which is helpful for users who are dedicated to virus knowledge mining.
ViLiD construction
Data source and dataset construction
The NCBI E-utils tool was used to obtain literature data, which was preprocessed to remove duplicates, non-English literature, and to correct coding issues, and metadata for each literature was downloaded in Medline format (containing title and abstract). Among them, the unlabeled raw text as of March 2, 2023 (12 viruses with a total of xxx abstracts) was used as the language modeling dataset. For virus topic classification, we focus on the existing coronavirus literature databases, which also contain the large number of papers. LitCovid and CoronalCentral provide two levels of topic classification, where the LitCovid classification is relatively broad, covering 7 main topics, namely “Mechanism”, “Transmission”, “Diagnosis”, “Treatment”, “Prevention”, “Case Report”, and “Epidemic Forecasting”, while CoronaCentral covers 32 more detailed topics, namely “Clinical Reports”, “Clinical Trials”, “Communication”, “Contact Tracing”, “Diagnostics”, “Drug Targets”, “Education”, “Effect on Medical Specialties”, “Forecasting & Modelling”, “Health Policy”, “Healthcare Workers”, “Imaging”, “Immunology”, “Inequality”, “Infection Reports”, “Long Haul”, “Medical Devices”, “Misinformation”, “Model Systems & Tools”, “Molecular Biology”, “Non-human”, “Non-medical”, “Pediatrics”, “Prevalence”, “Prevention”, “Psychology”, “Recommendations”, “Risk Factors”, “Surveillance”, “Therapeutics”, “Transmission”, and “Vaccines”. Therefore, we downloaded the labeled data from both databases as of February 20, 2023 and constructed 2 multi-label classification datasets. Since the two topic classification datasets are from coronaviruses, we manually annotated two external test sets from 8 disease-related virus literature to test the effect of model migration on other viruses. In addition to common article topics of virus literature, we are also interested in other information, including literature containing descriptions of viral drug resistance mutations, immune escape mutations, and viral integration site information, so we integrated literature from ViMIC, VISDB, and ViMRT databases as references to manually annotate the label of drug resistance mutation (DRM), immune escape mutation (IEM) and viral integration site (VIS) as the third dataset. The above three multi-label classification datasets were divided into training and validation sets in a ratio of 8:2. For the genomic data of the papers, we crawled all ClinVar, GEO, Protein, Nucleotide, and SRA information associated with each article and the links to each dataset through Entrez. To assess the indicators of attention of each article, we also crawled the Altmetric data associated with literature citations and attention as a reference for article visualization.
Multi-topic model construction
Since the category labels of LitCovid articles were assigned by human assessment as of June 2023, to evaluate existing language models on the multi-label classification task of virus literature, we fine-tuned different model architecture types and domain pre-trained models on the LitCovid dataset for comparison. The pre-trained model architectures include BERT, ELECTRA, ALBERT, RoBERTa, XLNet, and DistilBERT. In addition to the original pre-trained models of the 6 model architectures, we also fine-tuned 8 models derived from them in the biomedical domain, including BioBERT, BioClinicalBERT, BioLinkBERT, PubMedBERT, BioM-ELECTRA, PubMedELECTRA, BioM-ALBERT, and BioMed-RoBERTa. For the hyperparameters of the models, the learning rate was set to 2e-5, the batch size was set to 64, and each model was run for 5 epochs. Since most of the abstracts in the dataset have text lengths (title + abstract) less than 512 and the input sequence limit for most pre-trained language models is 512, we set max_seq_length to 512. Language modeling is well suited for tasks that require a good contextual understanding of the entire text sequence, which allows models to be adapted to the virus domain-specific corpus. To further improve the semantic understanding of the virus literature and the classification performance of the pre-trained language models, we proposed the virus-specific pre-training strategy, where the best-performing model is further pre-trained with masked language modeling on the virus literature corpus (unlabeled raw text), and the sliding window technique is used to optimize for long text. For the modeling configuration, the ratio of tokens to mask for MLM loss is 0.15 and the fraction for stride of the sliding window is 0.8. We run 100 epochs with a batch size of 150, then the virus-specific pre-training model is produced, which is also fine-tuned and tested on the three multi-label datasets and applied in downstream large-scale literature prediction, respectively. For the language modeling and downstream fine-tuning phases, we experimented on RTX 3090 and Tesla P100, respectively, using cross-entropy loss as the loss function and AdamW as the optimizer. These pre-trained language models were implemented using PyTorch (version 1.12.0) and Hugging Face Transformers (version 4.26.1).
Other information extraction and text generation
In order to target documents of interest by qualifying key entity information, we extracted entities from the literature, including general biomedical entities, viral domain entities, and cistrome factors. For generic biomedical entities, we use PubTator to identify “Gene”, “Disease”, “Chemical”, “Cell line”, “Species”, “DNA mutation”, “Protein mutation”, “SNP” with the unique normalized ID. In addition, we manually summarized and supplemented the entities associated with the severe disease-related virus based on literature research and CoronaCentral, and identified 8 categories of entities, including “Virus Genotype”, “Test Type”, “Transmission Type”, “Vaccine Type”, “Risk Factor”, “Prevention Method”, “Symptom”, “Medical Specialty”. Moreover, virus infection and integration have potential effects on the human genome, so we also identified cistrome factors (“Histone Modification”, “Transcription Factor Binding Site”, “Chromatin Accessibility”) from the literature. The virus-specific entities and cistrome factors were obtained by the custom rule-based approach. After entity recognition, we also identified the relations between entities using our biomedical relation extraction model which can identify 124 relations. Since journals have different requirements for article abstracts, resulting in many long and complex abstracts. To enable users to understand the main idea of the article more easily and quickly, we used an open-source text generation model Scientific Abstract Simplification (SAS) to rewrite the abstracts with more than 250 words to summarize the key points of the literature. We also evaluate the reproducibility of the SAS model by fine-tuning the rewritten text on the LC multi-label classification task.
Web interface
The web interface was built using the LNMP (Linix+Nginx+MySQL+PHP) framework. The use of Ajax makes JS execute asynchronous network requests, thereby reducing the need to reload all the content that the user is interested in each time. The front-end visual interaction is implemented using Apache ECharts, and the article list is integrated with a download function to provide users with literature of interest.
Contact
Zhang, Xiaoyan
Department of Bioinformatics, School of Life Sciences and Technology
Tongji University, No.1239, Siping Road, Shanghai, P.R. China
Email: xyzhang@tongji.edu.com
Zhang, Zeyu
PhD candidate
Department of Bioinformatics, School of Life Sciences and Technology
Tongji University, No.1239, Siping Road, Shanghai, P.R. China
Email: zhzyvv@gmail.com