ViMRT: a text-mining tool and search engine for automated virus mutation recognition
 Highlights(Docs)
Highlights(Docs)
 Viral mutation dataset construction
Viral mutation dataset construction
This study constructed a viral mutation dataset containing 2,492 abstracts on 5 viruses including HBV, HIV, HPV, HTLV1, and EBV serving for tool development. This corpus covered most specific written forms of viral mutation from different literatures.
 Virus mutation recognition tool
Virus mutation recognition tool
We proposed ViMRT, a text-mining tool for automated virus mutation recognition by developing 8 rule patterns and 12 regular expression patterns for different written forms of viral mutation based on natural language processing. Our tool outperforms other conventional tools like tmVar, nala and MutationFinder developed based on human corpus.
 The ViMRT web search engine
The ViMRT web search engine
A novel convenient search engine for retrieving mutation-specific information of 7 viruses that included virus gene and related human disease with co-occurrence relationship is provided, which can contribute to other downstream tasks, such as virus mutation database development, immune escape mechanism exploration, vaccine design and update, drug resistance mutations analysis and assistance of clinical personalized medicine.
 Method overview
Method overview
As shown in below Figure, ViMRT Firstly ceated the gold standard file of virus mutation from our previously developed ViMIC database, consisting of HBV, HIV, HPV, HTLV1 and EBV. Next, ViMRT mainly built 8 rule patterns to optimize and standardize mutation identification of tmVar and 12 regular expressions to recognize virus mutation from original literature. Finally, ViMRT created virus gene corpus from NCBI Pubmed and gene database and disease corpus from CTD database to identify genes and diseases, and developed a search engine using the Django 3.2.6 from virus mutation sentence.
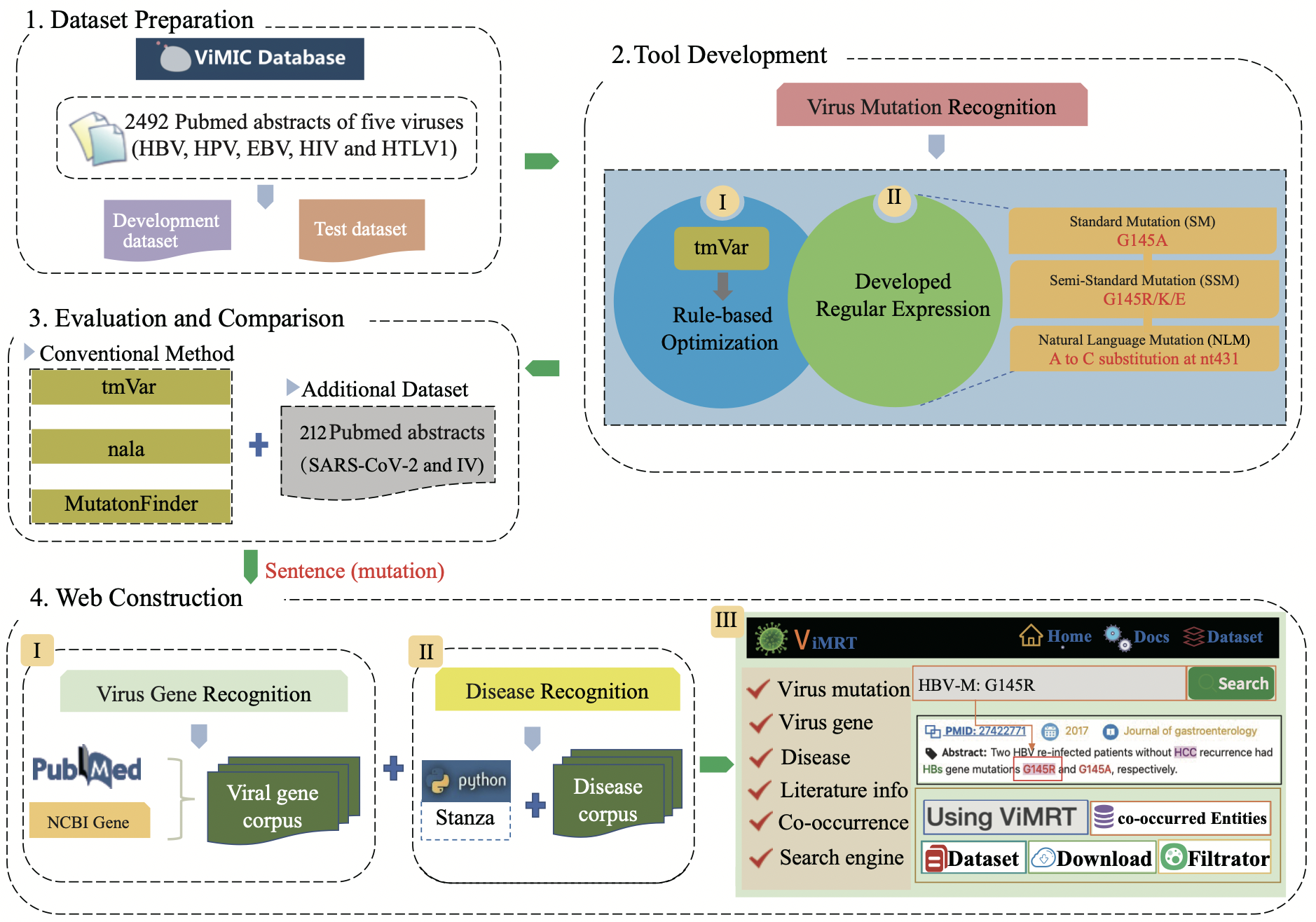
A schematic overview of the ViMRT project
 Dataset
Dataset
Table 1. Statistics of mutation corpus
| Dataset | Positive abstracts | Negative abstracts |
| Development set | 415 | 415 |
| Test set | 831 | 831 |
 Result
Result| Method | TP | FP | FN | Precision | Recall | F-score |
| ViMRT | 6058 | 37 | 64 | 99.39% | 98.95% | 99.17% |
| tmVar | 4230 | 393 | 1892 | 91.50% | 69.10% | 78.74% |
| Nala | 4212 | 278 | 1910 | 93.81% | 68.80% | 79.38% |
| MutationFinder | 3628 | 7 | 2494 | 99.81% | 59.26% | 74.37% |
 Download
DownloadI. Users can download the software:
ViMRT Software
ViMRT Corpus
II. Users can download mutation files of 7 viruses (update time:2022-05-06)
| Virus name | Entities | Data available |
| Hepatitis B Virus (HBV) | 13916 | Download |
| Human Papillomavirus Virus (HPV) | 2241 | Download |
| Epstein-Barr Virus (EBV) | 917 | Download |
| Human Immunodeficiency Virus (HIV) | 45686 | Download |
| Human T-cell Lymphotropic Virus type 1 (HTLV1) | 257 | Download |
| Influenza Virus (IV) | 16537 | Download |
| Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) | 40639 | Download |
